Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir
- PMID: 18597780
- PMCID: PMC2754059
- DOI: 10.1016/j.jmb.2008.05.062
Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir
Abstract
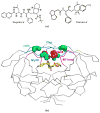
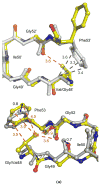
HIV-1 (human immunodeficiency virus type 1) protease (PR) and its mutants are important antiviral drug targets. The PR flap region is critical for binding substrates or inhibitors and catalytic activity. Hence, mutations of flap residues frequently contribute to reduced susceptibility to PR inhibitors in drug-resistant HIV. Structural and kinetic analyses were used to investigate the role of flap residues Gly48, Ile50, and Ile54 in the development of drug resistance. The crystal structures of flap mutants PR(I50V) (PR with I50V mutation), PR(I54V) (PR with I54V mutation), and PR(I54M) (PR with I54M mutation) complexed with saquinavir (SQV) as well as PR(G48V) (PR with G48V mutation), PR(I54V), and PR(I54M) complexed with darunavir (DRV) were determined at resolutions of 1.05-1.40 A. The PR mutants showed changes in flap conformation, interactions with adjacent residues, inhibitor binding, and the conformation of the 80s loop relative to the wild-type PR. The PR contacts with DRV were closer in PR(G48V)-DRV than in the wild-type PR-DRV, whereas they were longer in PR(I54M)-DRV. The relative inhibition of PR(I54V) and that of PR(I54M) were similar for SQV and DRV. PR(G48V) was about twofold less susceptible to SQV than to DRV, whereas the opposite was observed for PR(I50V). The observed inhibition was in agreement with the association of G48V and I50V with clinical resistance to SQV and DRV, respectively. This analysis of structural and kinetic effects of the mutants will assist in the development of more effective inhibitors for drug-resistant HIV.
Figures

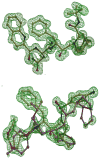
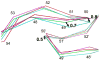



Similar articles
-
Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters.FEBS J. 2010 Sep;277(18):3699-714. doi: 10.1111/j.1742-4658.2010.07771.x. Epub 2010 Aug 2. FEBS J. 2010. PMID: 20695887 Free PMC article.
-
Structural and kinetic analyses of the protease from an amprenavir-resistant human immunodeficiency virus type 1 mutant rendered resistant to saquinavir and resensitized to amprenavir.J Virol. 2000 Aug;74(16):7636-41. doi: 10.1128/jvi.74.16.7636-7641.2000. J Virol. 2000. PMID: 10906218 Free PMC article.
-
Systematic molecular dynamics, MM-PBSA, and ab initio approaches to the saquinavir resistance mechanism in HIV-1 PR due to 11 double and multiple mutations.J Phys Chem B. 2014 Aug 14;118(32):9538-52. doi: 10.1021/jp502687q. Epub 2014 Jul 31. J Phys Chem B. 2014. PMID: 25036111
-
[Resistance to darunavir].Enferm Infecc Microbiol Clin. 2008 Oct;26 Suppl 10:51-60. doi: 10.1016/s0213-005x(08)76554-6. Enferm Infecc Microbiol Clin. 2008. PMID: 19195460 Review. Spanish.
-
Resilience to resistance of HIV-1 protease inhibitors: profile of darunavir.AIDS Rev. 2008 Jul-Sep;10(3):131-42. AIDS Rev. 2008. PMID: 18820715 Free PMC article. Review.
Cited by
-
Structural Studies of a Rationally Selected Multi-Drug Resistant HIV-1 Protease Reveal Synergistic Effect of Distal Mutations on Flap Dynamics.PLoS One. 2016 Dec 16;11(12):e0168616. doi: 10.1371/journal.pone.0168616. eCollection 2016. PLoS One. 2016. PMID: 27992544 Free PMC article.
-
Impacts of drug resistance mutations on the structural asymmetry of the HIV-2 protease.BMC Mol Cell Biol. 2020 Jun 23;21(1):46. doi: 10.1186/s12860-020-00290-1. BMC Mol Cell Biol. 2020. PMID: 32576133 Free PMC article.
-
Drug Resistance Mechanism of M46I-Mutation-Induced Saquinavir Resistance in HIV-1 Protease Using Molecular Dynamics Simulation and Binding Energy Calculation.Viruses. 2022 Mar 28;14(4):697. doi: 10.3390/v14040697. Viruses. 2022. PMID: 35458427 Free PMC article.
-
Highly drug-resistant HIV-1 protease reveals decreased intra-subunit interactions due to clusters of mutations.FEBS J. 2020 Aug;287(15):3235-3254. doi: 10.1111/febs.15207. Epub 2020 Jan 23. FEBS J. 2020. PMID: 31920003 Free PMC article.
-
Novel P2 tris-tetrahydrofuran group in antiviral compound 1 (GRL-0519) fills the S2 binding pocket of selected mutants of HIV-1 protease.J Med Chem. 2013 Feb 14;56(3):1074-83. doi: 10.1021/jm301519z. Epub 2013 Jan 23. J Med Chem. 2013. PMID: 23298236 Free PMC article.
References
-
- Miller M, Schneider J, Sathyanarayana BK, Toth MV, Marshall GR, Clawson L, Selk L, Kent SB, Wlodawer A. Structure of complex of synthetic HIV-1 protease with a substrate-based inhibitor at 2.3 A resolution. Science. 1989;246:1149–52. - PubMed
-
- Gustchina A, Weber IT. Comparison of inhibitor binding in HIV-1 protease and in non-viral aspartic proteases: the role of the flap. FEBS. 1990;269:269–272. - PubMed
-
- Ishima R, Freedberg DI, Wang YX, Louis JM, Torchia DA. Flap opening and dimer-interface flexibility in the free and inhibitor-bound HIV protease. Structure. 1999;7:1047–55. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

