The AID/APOBEC family of nucleic acid mutators
- PMID: 18598372
- PMCID: PMC2481415
- DOI: 10.1186/gb-2008-9-6-229
The AID/APOBEC family of nucleic acid mutators
Abstract
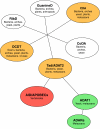
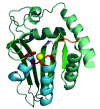
The AID/APOBECs, a group of cytidine deaminases, represent a somewhat unusual protein family that can insert mutations in DNA and RNA as a result of their ability to deaminate cytidine to uridine. The ancestral AID/APOBECs originated from a branch of the zinc-dependent deaminase superfamily at the beginning of the vertebrate radiation. Other members of the family have arisen in mammals and present a history of complex gene duplications and positive selection. All AID/APOBECs have a characteristic zinc-coordination motif, which forms the core of the catalytic site. The crystal structure of human APOBEC2 shows remarkable similarities to that of the bacterial tRNA-editing enzyme TadA, which suggests a conserved mechanism by which polynucleotides are recognized and deaminated. The AID/APOBECs seem to have diverse roles. AID and the APOBEC3s are DNA mutators, acting in antigen-driven antibody diversification processes and in an innate defense system against retroviruses, respectively. APOBEC1 edits the mRNA for apolipoprotein B, a protein involved in lipid transport. A detailed understanding of the biological roles of the family is still some way off, however, and the functions of some members of the family are completely unknown. Given their ability to mutate DNA, a role for the AID/APOBECs in the onset of cancer has been proposed.
Figures

References
-
- Navaratnam N, Morrison JR, Bhattacharya S, Patel D, Funahashi T, Giannoni F, Teng BB, Davidson NO, Scott J. The p27 catalytic subunit of the apolipoprotein B mRNA editing enzyme is a cytidine deaminase. J Biol Chem. 1993;268:20709–20712. - PubMed
-
- Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

