Circular reasoning rather than cyclic expression
- PMID: 18598377
- PMCID: PMC2481420
- DOI: 10.1186/gb-2008-9-6-403
Circular reasoning rather than cyclic expression
Abstract
A response to Combined analysis reveals a core set of cycling genes by Y Lu, S Mahony, PV Benos, R Rosenfeld, I Simon, LL Breeden and Z Bar-Joseph. Genome Biol 2007, 8:R146.
Figures
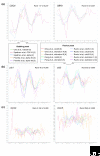

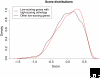

Comment on
-
Combined analysis reveals a core set of cycling genes.Genome Biol. 2007;8(7):R146. doi: 10.1186/gb-2007-8-7-r146. Genome Biol. 2007. PMID: 17650318 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

