Application of metabolomics to cardiovascular biomarker and pathway discovery
- PMID: 18598890
- PMCID: PMC3204897
- DOI: 10.1016/j.jacc.2008.03.043
Application of metabolomics to cardiovascular biomarker and pathway discovery
Abstract
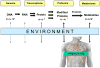
Emerging technologies based on mass spectrometry and nuclear magnetic resonance enable the monitoring of hundreds of metabolites from tissues or body fluids, that is, "metabolomics." Because metabolites change rapidly in response to physiologic perturbations, they represent proximal reporters of disease phenotypes. The profiling of low molecular weight biochemicals, including lipids, sugars, nucleotides, organic acids, and amino acids, that serve as substrates and products in metabolic pathways is particularly relevant to cardiovascular diseases. In addition to serving as disease biomarkers, circulating metabolites may participate in previously unanticipated roles as regulatory signals with hormone-like functions. Cellular metabolic pathways are highly conserved among species, facilitating complementary functional studies in model organisms to provide insight into metabolic changes identified in humans. Although metabolic profiling technologies and methods of pattern recognition and data reduction remain under development, the coupling of metabolomics with other functional genomic approaches promises to extend our ability to elucidate biological pathways and discover biomarkers of human disease.
Conflict of interest statement
There are no potential conflicts of interest or financial disclosures for this manuscript.
Figures
References
-
- Horning EC, Horning MG. Metabolic profiles: gas-phase methods for analysis of metabolites. Clin Chem. 1971;17:802–9. - PubMed
-
- Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181–9. - PubMed
-
- Raamsdonk LM, Teusink B, Broadhurst D, et al. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat Biotechnol. 2001;19:45–50. - PubMed
-
- Allen J, Davey HM, Broadhurst D, et al. High-throughput classification of yeast mutants for functional genomics using metabolic footprinting. Nat Biotechnol. 2003;21:692–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

