Mutations in Fbx4 inhibit dimerization of the SCF(Fbx4) ligase and contribute to cyclin D1 overexpression in human cancer
- PMID: 18598945
- PMCID: PMC2597358
- DOI: 10.1016/j.ccr.2008.05.017
Mutations in Fbx4 inhibit dimerization of the SCF(Fbx4) ligase and contribute to cyclin D1 overexpression in human cancer
Abstract
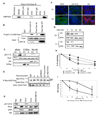
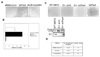
SCF(Fbx4) was recently identified as the E3 ligase for cyclin D1. We now describe cell-cycle-dependent phosphorylation and dimerization of Fbx4 that is regulated by GSK3beta and is defective in human cancer. We present data demonstrating that a pathway involving Ras-Akt-GSK3beta controls the temporal phosphorylation and dimerization of the SCF(Fbx4) E3 ligase. Inhibition of Fbx4 activity results in accumulation of nuclear cyclin D1 and oncogenic transformation. The importance of this regulatory pathway for normal cell growth is emphasized by the prevalence of mutations in Fbx4 in human cancer that impair dimerization. Collectively, these data reveal that inactivation of the cyclin D1 E3 ligase likely contributes to cyclin D1 overexpression in a significant fraction of human cancer.
Figures




References
-
- Aggarwal P, Lessie MD, Lin DI, Pontano L, Gladden AB, Nuskey B, Goradia A, Wasik MA, Klein-Szanto AJ, Rustgi AK, et al. Nuclear accumulation of cyclin D1 during S phase inhibits Cul4-dependent Cdt1 proteolysis and triggers p53-dependent DNA rereplication. Genes Dev. 2007;21:2908–2922. - PMC - PubMed
-
- Benzeno S, L F, Guo M, Barbash O, Zhang F, Herman JG, Klein PS, Rustgi A, Diehl JA. Identification of mutations that disrupt phosphorylation-dependent nuclear export of cyclin D1. Oncogene. 2006 - PubMed
-
- Chew EH, Poobalasingam T, Hawkey CJ, Hagen T. Characterization of cullin-based E3 ubiquitin ligases in intact mammalian cells--evidence for cullin dimerization. Cell Signal. 2007;19:1071–1080. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

