Structure of IL-22 bound to its high-affinity IL-22R1 chain
- PMID: 18599299
- PMCID: PMC2637415
- DOI: 10.1016/j.str.2008.06.005
Structure of IL-22 bound to its high-affinity IL-22R1 chain
Abstract
IL-22 is an IL-10 family cytokine that initiates innate immune responses against bacterial pathogens and contributes to immune disease. IL-22 biological activity is initiated by binding to a cell-surface complex composed of IL-22R1 and IL-10R2 receptor chains and further regulated by interactions with a soluble binding protein, IL-22BP, which shares sequence similarity with an extracellular region of IL-22R1 (sIL-22R1). IL-22R1 also pairs with the IL-20R2 chain to induce IL-20 and IL-24 signaling. To define the molecular basis of these diverse interactions, we have determined the structure of the IL-22/sIL-22R1 complex. The structure, combined with homology modeling and surface plasmon resonance studies, defines the molecular basis for the distinct affinities and specificities of IL-22 and IL-10 receptor chains that regulate cellular targeting and signal transduction to elicit effective immune responses.
Figures


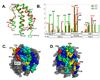



References
-
- Blumberg H, Conklin D, Xu WF, Grossmann A, Brender T, Carollo S, Eagan M, Foster D, Haldeman BA, Hammond A, et al. Interleukin 20: discovery, receptor identification, and role in epidermal function. Cell. 2001;104:9–19. - PubMed
-
- Brown RJ, Adams JJ, Pelekanos RA, Wan Y, McKinstry WJ, Palethorpe K, Seeber RM, Monks TA, Eidne KA, Parker MW, Waters MJ. Model for growth hormone receptor activation based on subunit rotation within a receptor dimer. Nat Struct Mol Biol. 2005;12:814–821. - PubMed
-
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–921. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

