The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport
- PMID: 18599740
- PMCID: PMC3654663
- DOI: 10.1126/science.1160406
The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport
Abstract
Membrane transporters that use energy stored in sodium gradients to drive nutrients into cells constitute a major class of proteins. We report the crystal structure of a member of the solute sodium symporters (SSS), the Vibrio parahaemolyticus sodium/galactose symporter (vSGLT). The approximately 3.0 angstrom structure contains 14 transmembrane (TM) helices in an inward-facing conformation with a core structure of inverted repeats of 5 TM helices (TM2 to TM6 and TM7 to TM11). Galactose is bound in the center of the core, occluded from the outside solutions by hydrophobic residues. Surprisingly, the architecture of the core is similar to that of the leucine transporter (LeuT) from a different gene family. Modeling the outward-facing conformation based on the LeuT structure, in conjunction with biophysical data, provides insight into structural rearrangements for active transport.
Figures









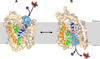
Comment in
-
Structural biology. Symmetric transporters for asymmetric transport.Science. 2008 Aug 8;321(5890):781-2. doi: 10.1126/science.1161495. Science. 2008. PMID: 18687947 Free PMC article. No abstract available.
References
-
- Crane R, Miller D, Bihler I. In: Membrane Transport and Metabolism. Kleinzeller A, Kotyk A, editors. London: Academic Press; 1961. pp. 439–449.
-
- Schultz SG, Curran PF. Physiol Rev. 1970 Oct;50:637. - PubMed
-
- Wright EM, Loo DD, Hirayama BA, Turk E. Physiology (Bethesda) 2004 Dec;19:370. - PubMed
-
- Wright EM, Hirayama BA, Loo DF. J Intern Med. 2007 Jan;261:32. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

