Robust, tunable biological oscillations from interlinked positive and negative feedback loops
- PMID: 18599789
- PMCID: PMC2728800
- DOI: 10.1126/science.1156951
Robust, tunable biological oscillations from interlinked positive and negative feedback loops
Abstract
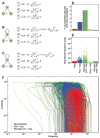
A simple negative feedback loop of interacting genes or proteins has the potential to generate sustained oscillations. However, many biological oscillators also have a positive feedback loop, raising the question of what advantages the extra loop imparts. Through computational studies, we show that it is generally difficult to adjust a negative feedback oscillator's frequency without compromising its amplitude, whereas with positive-plus-negative feedback, one can achieve a widely tunable frequency and near-constant amplitude. This tunability makes the latter design suitable for biological rhythms like heartbeats and cell cycles that need to provide a constant output over a range of frequencies. Positive-plus-negative oscillators also appear to be more robust and easier to evolve, rationalizing why they are found in contexts where an adjustable frequency is unimportant.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

