The Yku70-Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle
- PMID: 18600234
- PMCID: PMC2515202
- DOI: 10.1038/embor.2008.121
The Yku70-Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle
Abstract
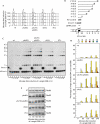
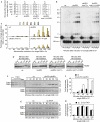
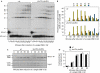
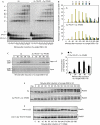
DNA double-strand breaks (DSBs) are repaired by non-homologous end joining (NHEJ) or homologous recombination (HR). HR requires 5' DSB end degradation that occurs in the presence of cyclin-dependent kinase (CDK) activity. Here, we show that a lack of any of the NHEJ proteins Yku (Yku70-Yku80), Lif1 or DNA ligase IV (Dnl4) increases 5' DSB end degradation in G1 phase, with ykuDelta cells showing the strongest effect. This increase depends on MRX, the recruitment of which at DSBs is enhanced in ykuDelta G1 cells. DSB processing in G2 is not influenced by the absence of Yku, but it is delayed by Yku overproduction, which also decreases MRX loading on DSBs. Moreover, DSB resection in ykuDelta cells occurs independently of CDK activity, suggesting that it might be promoted by CDK-dependent inhibition of Yku.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Daley JM, Palmbos PL, Wu D, Wilson TE (2005) Nonhomologous end joining in yeast. Annu Rev Genet 39: 431–451 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

