A high-resolution radiation hybrid map of rhesus macaque chromosome 5 identifies rearrangements in the genome assembly
- PMID: 18601997
- PMCID: PMC2605074
- DOI: 10.1016/j.ygeno.2008.05.013
A high-resolution radiation hybrid map of rhesus macaque chromosome 5 identifies rearrangements in the genome assembly
Abstract
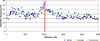
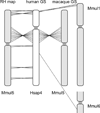
A 10,000-rad radiation hybrid (RH) cell panel of the rhesus macaque was generated to construct a comprehensive RH map of chromosome 5. The map represents 218 markers typed in 185 RH clones. The 4846-cR map has an average marker spacing of 798 kb. Alignments of the RH map to macaque and human genome sequences confirm a large inversion and reveal a previously unreported telomeric inversion. The macaque genome sequence indicates small translocations from the ancestral homolog of macaque chromosome 5 to macaque chromosomes 1 and 6. The RH map suggests that these are probably assembly artifacts. Unlike the genome sequence, the RH mapping data indicate the conservation of synteny between macaque chromosome 5 and human chromosome 4. This study shows that the 10,000-rad panel is appropriate for the generation of a high-resolution whole-genome RH map suitable for the verification of the rhesus genome assembly.
Figures





References
-
- Gibbs RA, et al. Evolutionary and biomedical insights from the rhesus macaque genome. Science. 2007;316:222–234. - PubMed
-
- Olivier M, et al. A high-resolution radiation hybrid map of the human genome draft sequence. Science. 2001;291:1298–1302. - PubMed
-
- Weikard R, Goldammer T, Laurent P, Womack JE, Kuehn C. A gene-based high-resolution comparative radiation hybrid map as a framework for genome sequence assembly of a bovine chromosome 6 region associated with QTL for growth, body composition, and milk performance traits. BMC Genomics. 2006;7:53. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

