SecA, the motor of the secretion machine, binds diverse partners on one interactive surface
- PMID: 18602400
- PMCID: PMC2633600
- DOI: 10.1016/j.jmb.2008.06.049
SecA, the motor of the secretion machine, binds diverse partners on one interactive surface
Abstract
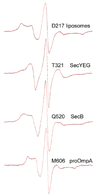
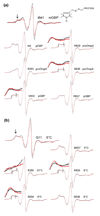
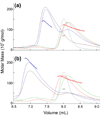
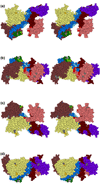
In all living cells, regulated passage across membranes of specific proteins occurs through a universally conserved secretory channel. In bacteria and chloroplasts, the energy for the mechanical work of moving polypeptides through that channel is provided by SecA, a regulated ATPase. Here, we use site-directed spin labeling and electron paramagnetic resonance spectroscopy to identify the interactive surface used by SecA for each of the diverse binding partners encountered during the dynamic cycle of export. Although the binding sites overlap, resolution at the level of aminoacyl side chains allows us to identify contacts that are unique to each partner. Patterns of constraint and mobilization of residues on that interactive surface suggest a conformational change that may underlie the coupling of ATP hydrolysis to precursor translocation.
Figures







References
-
- Papanikou E, Karamanou S, Economou A. Bacterial protein secretion through the translocase nanomachine. Nat. Rev. Microbiol. 2007;5:839–851. - PubMed
-
- Crane JM, Mao C, Lilly AA, Smith VF, Suo Y, Hubbell WL, Randall LL. Mapping of the docking of SecA onto the chaperone SecB by site-directed spin labeling: insight into the mechanism of ligand transfer during protein export. J. Mol. Biol. 2005;353:295–307. - PubMed
-
- Zimmer J, Li W, Rapoport TA. A novel dimer interface and conformational changes revealed by an X-ray structure of B. subtilis SecA. J. Mol. Biol. 2006;364:259–265. - PubMed
-
- Hunt JF, Weinkauf S, Henry L, Fak JJ, McNicholas P, Oliver DB, Deisenhofer J. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science. 2002;297:2018–2026. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

