Chemical mechanisms of histone lysine and arginine modifications
- PMID: 18603028
- PMCID: PMC2642981
- DOI: 10.1016/j.bbagrm.2008.06.005
Chemical mechanisms of histone lysine and arginine modifications
Abstract
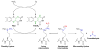
Histone lysine and arginine residues are subject to a wide array of post-translational modifications including methylation, citrullination, acetylation, ubiquitination, and sumoylation. The combinatorial action of these modifications regulates critical DNA processes including replication, repair, and transcription. In addition, enzymes that modify histone lysine and arginine residues have been correlated with a variety of human diseases including arthritis, cancer, heart disease, diabetes, and neurodegenerative disorders. Thus, it is important to fully understand the detailed kinetic and chemical mechanisms of these enzymes. Here, we review recent progress towards determining the mechanisms of histone lysine and arginine modifying enzymes. In particular, the mechanisms of S-adenosyl-methionine (AdoMet) dependent methyltransferases, FAD-dependent demethylases, iron dependent demethylases, acetyl-CoA dependent acetyltransferases, zinc dependent deacetylases, NAD(+) dependent deacetylases, and protein arginine deiminases are covered. Particular attention is paid to the conserved active-site residues necessary for catalysis and the individual chemical steps along the catalytic pathway. When appropriate, areas requiring further work are discussed.
Figures






References
-
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–94. - PubMed
-
- Luger K, Hansen JC. Nucleosome and chromatin fiber dynamics. Curr Opin Struct Biol. 2005;15:188–96. - PubMed
-
- Peterson CL, Laniel MA. Histones and histone modifications. Curr Biol. 2004;14:R546–51. - PubMed
-
- Suzuki A, Yamada R, Yamamoto K. Citrullination by peptidylarginine deiminase in rheumatoid arthritis. Ann N Y Acad Sci. 2007;1108:323–39. - PubMed
-
- Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26:5420–32. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

