Regulation of TORC1 by Rag GTPases in nutrient response
- PMID: 18604198
- PMCID: PMC2711503
- DOI: 10.1038/ncb1753
Regulation of TORC1 by Rag GTPases in nutrient response
Abstract
TORC1 (target of rapamycin complex 1) has a crucial role in the regulation of cell growth and size. A wide range of signals, including amino acids, is known to activate TORC1. Here, we report the identification of Rag GTPases as activators of TORC1 in response to amino acid signals. Knockdown of Rag gene expression suppressed the stimulatory effect of amino acids on TORC1 in Drosophila melanogaster S2 cells. Expression of constitutively active (GTP-bound) Rag in mammalian cells activated TORC1 in the absence of amino acids, whereas expression of dominant-negative Rag blocked the stimulatory effects of amino acids on TORC1. Genetic studies in Drosophila also show that Rag GTPases regulate cell growth, autophagy and animal viability during starvation. Our studies establish a function of Rag GTPases in TORC1 activation in response to amino acid signals.
Figures

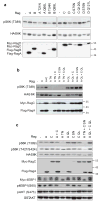

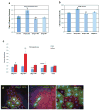

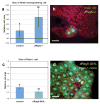
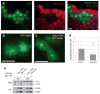
Comment in
-
Nutrient sensing: TOR's Ragtime.Nat Cell Biol. 2008 Aug;10(8):881-3. doi: 10.1038/ncb0808-881. Nat Cell Biol. 2008. PMID: 18670446 No abstract available.
References
-
- Hay N, Sonenberg N. Upstream and down stream of mTOR. Genes Dev. 2004;18:1926–1945. - PubMed
-
- Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. - PubMed
-
- Sabatini DM, Erdjument-Bromage H, Lui M, Tempst P, Snyder SH. RAFT1: a mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell. 1994;78:35–43. - PubMed
-
- Jacinto E, et al. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–1128. - PubMed
-
- Loewith R, et al. Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell. 2002;10:457–468. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

