Estimation of the available free energy in a LOV2-J alpha photoswitch
- PMID: 18604202
- PMCID: PMC2597337
- DOI: 10.1038/nchembio.99
Estimation of the available free energy in a LOV2-J alpha photoswitch
Abstract
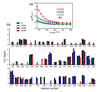
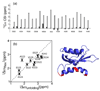
Protein photosensors are versatile tools for studying ligand-regulated allostery and signaling. Fundamental to these processes is the amount of energy that can be provided by a photosensor to control downstream signaling events. Such regulation is exemplified by the phototropins--plant serine/threonine kinases that are activated by blue light via conserved LOV (light, oxygen and voltage) domains. The core photosensor of oat phototropin 1 is a LOV domain that interacts in a light-dependent fashion with an adjacent alpha-helix (J alpha) to control kinase activity. We used solution NMR measurements to quantify the free energy of the LOV domain-J alpha-helix binding equilibrium in the dark and lit states. These data indicate that light shifts this equilibrium by approximately 3.8 kcal mol(-1), thus quantifying the energy available through LOV-J alpha for light-driven allosteric regulation. This study provides insight into the energetics of light sensing by phototropins and benchmark values for engineering photoswitchable systems based on the LOV-J alpha interaction.
Figures



Comment in
-
Protein dynamics under light control.Nat Chem Biol. 2008 Aug;4(8):449-50. doi: 10.1038/nchembio0808-449. Nat Chem Biol. 2008. PMID: 18641621
References
-
- Bhattacharyya RP, Remenyi A, Yeh BJ, Lim WA. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu Rev Biochem. 2006;75:655–680. - PubMed
-
- Pawson T. Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. 2004;116:191–203. - PubMed
-
- van der Horst MA, Hellingwerf KJ. Photoreceptor proteins, "Star Actors of Modern Times": A review of the functional dynamics in the structure of representative members of six different photoreceptor families. Acc Chem Res. 2004;37:13–20. - PubMed
-
- Christie JM. Phototropin blue-light receptors. Annu Rev Plant Biol. 2007;58:21–45. - PubMed
-
- Huala E, et al. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science. 1997;278:2120–2123. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

