Application of pressurized solvents for ultrafast trypsin hydrolysis in proteomics: proteomics on the fly
- PMID: 18605748
- PMCID: PMC2744211
- DOI: 10.1021/pr7008077
Application of pressurized solvents for ultrafast trypsin hydrolysis in proteomics: proteomics on the fly
Abstract
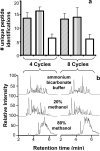
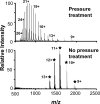
A new method for rapid proteolytic digestion of proteins under high pressure that uses pressure cycling technology in the range of 5-35 kpsi was demonstrated for proteomic analysis. Successful in-solution digestions of single proteins and complex protein mixtures were achieved in 60 s and then analyzed by reversed phase liquid chromatography-electrospray ionization ion trap-mass spectrometry. Method performance in terms of the number of Shewanella oneidensis peptides and proteins identified in a shotgun approach was evaluated relative to a traditional "overnight" sample preparation method. Advantages of the new method include greatly simplified sample processing, easy implementation, no cross contamination among samples, and cost effectiveness.
Figures
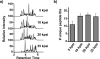

References
-
- Fischer HP. Towards quantitative biology: integration of biological information to elucidate disease pathways and to guide drug discovery. Biotechnol. Annu. Rev. 2005;11:1–68. - PubMed
-
- Hood L, Perlmutter RM. The impact of systems approaches on biological problems in drug discovery. Nat. Biotechnol. 2004;22(10):1215–7. - PubMed
-
- Blackler AR, Klammer AA, MacCoss MJ, Wu CC. Quantitative comparison of proteomic data quality between a 2D and 3D quadrupole ion trap. Anal. Chem. 2006;78(4):1337–44. - PubMed
-
- Denison C, Rudner AD, Gerber SA, Bakalarski CE, Moazed D, Gygi SP. A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol. Cell. Proteomics. 2005;4(3):246–54. - PubMed
-
- Hu Q, Noll RJ, Li H, Makarov A, Hardman M, Graham Cooks R. The Orbitrap: a new mass spectrometer. J. Mass. Spectrom. 2005;40(4):430–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

