The structural basis for peptidomimetic inhibition of eukaryotic ribonucleotide reductase: a conformationally flexible pharmacophore
- PMID: 18610997
- PMCID: PMC2726620
- DOI: 10.1021/jm800350u
The structural basis for peptidomimetic inhibition of eukaryotic ribonucleotide reductase: a conformationally flexible pharmacophore
Abstract
Eukaryotic ribonucleotide reductase (RR) catalyzes nucleoside diphosphate conversion to deoxynucleoside diphosphate. Crucial for rapidly dividing cells, RR is a target for cancer therapy. RR activity requires formation of a complex between subunits R1 and R2 in which the R2 C-terminal peptide binds to R1. Here we report crystal structures of heterocomplexes containing mammalian R2 C-terminal heptapeptide, P7 (Ac-1FTLDADF7) and its peptidomimetic P6 (1Fmoc(Me)PhgLDChaDF7) bound to Saccharomyces cerevisiae R1 (ScR1). P7 and P6, both of which inhibit ScRR, each bind at two contiguous sites containing residues that are highly conserved among eukaryotes. Such binding is quite distinct from that reported for prokaryotes. The Fmoc group in P6 peptide makes several hydrophobic interactions that contribute to its enhanced potency in binding to ScR1. Combining all of our results, we observe three distinct conformations for peptide binding to ScR1. These structures provide pharmacophores for designing highly potent nonpeptide class I RR inhibitors.
Figures

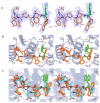
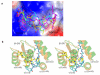

References
-
- Stubbe J. Ribonucleotide reductases: the link between an RNA and a DNA world? Curr Opin Struct Biol. 2000;10:731–6. - PubMed
-
- Bollinger JM, Jr., Edmondson DE, Huynh BH, Filley J, Norton JR, Stubbe J. Mechanism of assembly of the tyrosyl radical-dinuclear iron cluster cofactor of ribonucleotide reductase. Science. 1991;253:292–8. - PubMed
-
- Fontecave M, Eliasson R, Reichard P. Enzymatic regulation of the radical content of the small subunit of Escherichia coli ribonucleotide reductase involving reduction of its redox centers. J Biol Chem. 1989;264:9164–70. - PubMed
-
- Sommerhalter M, Voegtli WC, Perlstein DL, Ge J, Stubbe J, Rosenzweig AC. Structures of the yeast ribonucleotide reductase Rnr2 and Rnr4 homodimers. Biochemistry. 2004;43:7736–42. - PubMed
-
- van der Donk WA, Yu G, Silva DJ, Stubbe J, McCarthy JR, Jarvi ET, Matthews DP, Resvick RJ, Wagner E. Inactivation of ribonucleotide reductase by (E)-2′-fluoromethylene-2′-deoxycytidine 5′-diphosphate: a paradigm for nucleotide mechanism-based inhibitors. Biochemistry. 1996;35:8381–91. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

