Discriminatory accuracy from single-nucleotide polymorphisms in models to predict breast cancer risk
- PMID: 18612136
- PMCID: PMC2528005
- DOI: 10.1093/jnci/djn180
Discriminatory accuracy from single-nucleotide polymorphisms in models to predict breast cancer risk
Abstract
One purpose for seeking common alleles that are associated with disease is to use them to improve models for projecting individualized disease risk. Two genome-wide association studies and a study of candidate genes recently identified seven common single-nucleotide polymorphisms (SNPs) that were associated with breast cancer risk in independent samples. These seven SNPs were located in FGFR2, TNRC9 (now known as TOX3), MAP3K1, LSP1, CASP8, chromosomal region 8q, and chromosomal region 2q35. I used estimates of relative risks and allele frequencies from these studies to estimate how much these SNPs could improve discriminatory accuracy measured as the area under the receiver operating characteristic curve (AUC). A model with these seven SNPs (AUC = 0.574) and a hypothetical model with 14 such SNPs (AUC = 0.604) have less discriminatory accuracy than a model, the National Cancer Institute's Breast Cancer Risk Assessment Tool (BCRAT), that is based on ages at menarche and at first live birth, family history of breast cancer, and history of breast biopsy examinations (AUC = 0.607). Adding the seven SNPs to BCRAT improved discriminatory accuracy to an AUC of 0.632, which was, however, less than the improvement from adding mammographic density. Thus, these seven common alleles provide less discriminatory accuracy than BCRAT but have the potential to improve the discriminatory accuracy of BCRAT modestly. Experience to date and quantitative arguments indicate that a huge increase in the numbers of case patients with breast cancer and control subjects would be required in genome-wide association studies to find enough SNPs to achieve high discriminatory accuracy.
Figures
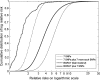
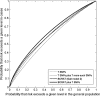
Comment in
-
Gauging the performance of SNPs, biomarkers, and clinical factors for predicting risk of breast cancer.J Natl Cancer Inst. 2008 Jul 16;100(14):978-9. doi: 10.1093/jnci/djn215. Epub 2008 Jul 8. J Natl Cancer Inst. 2008. PMID: 18612128 Free PMC article. No abstract available.
-
Re: Discriminatory accuracy from single-nucleotide polymorphisms in models to predict breast cancer risk.J Natl Cancer Inst. 2009 Dec 16;101(24):1731-2; author reply 1732. doi: 10.1093/jnci/djp394. J Natl Cancer Inst. 2009. PMID: 19903803 Free PMC article. No abstract available.
References
-
- Evans JP. Health care in the age of genetic medicine. JAMA. 2007;298(22):2670–2672. - PubMed
-
- Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat Genet. 2007;39(7):865–869. - PubMed
-
- Cox A, Dunning AM, Garcia-Closas M, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39(3):352–358. - PubMed
-
- Costantino JP, Gail MH, Pee D, et al. Validation studies for models projecting the risk of invasive and total breast cancer incidence. J Natl Cancer Inst. 1999;91(18):1541–1548. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

