A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis
- PMID: 18612301
- PMCID: PMC2644410
- DOI: 10.1038/nbt1414
A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis
Abstract
DNA methylation is an indispensible epigenetic modification required for regulating the expression of mammalian genomes. Immunoprecipitation-based methods for DNA methylome analysis are rapidly shifting the bottleneck in this field from data generation to data analysis, necessitating the development of better analytical tools. In particular, an inability to estimate absolute methylation levels remains a major analytical difficulty associated with immunoprecipitation-based DNA methylation profiling. To address this issue, we developed a cross-platform algorithm-Bayesian tool for methylation analysis (Batman)-for analyzing methylated DNA immunoprecipitation (MeDIP) profiles generated using oligonucleotide arrays (MeDIP-chip) or next-generation sequencing (MeDIP-seq). We developed the latter approach to provide a high-resolution whole-genome DNA methylation profile (DNA methylome) of a mammalian genome. Strong correlation of our data, obtained using mature human spermatozoa, with those obtained using bisulfite sequencing suggest that combining MeDIP-seq or MeDIP-chip with Batman provides a robust, quantitative and cost-effective functional genomic strategy for elucidating the function of DNA methylation.
Figures
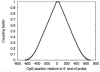

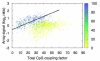




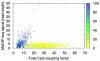

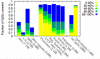

References
-
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. - PubMed
-
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed
-
- Beck S, Rakyan VK. The methylome: approaches for global DNA methylation profiling. Trends Genet. 2008;24:231–237. - PubMed
-
- Tompa R, et al. Genome-wide profiling of DNA methylation reveals transposon targets of CHROMOMETHYLASE3. Curr Biol. 2002;12:65–68. - PubMed
-
- Lippman Z, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

