The role of cardiac troponin T quantity and function in cardiac development and dilated cardiomyopathy
- PMID: 18612386
- PMCID: PMC2441440
- DOI: 10.1371/journal.pone.0002642
The role of cardiac troponin T quantity and function in cardiac development and dilated cardiomyopathy
Abstract
Background: Hypertrophic (HCM) and dilated (DCM) cardiomyopathies result from sarcomeric protein mutations, including cardiac troponin T (cTnT, TNNT2). We determined whether TNNT2 mutations cause cardiomyopathies by altering cTnT function or quantity; whether the severity of DCM is related to the ratio of mutant to wildtype cTnT; whether Ca(2+) desensitization occurs in DCM; and whether absence of cTnT impairs early embryonic cardiogenesis.
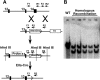
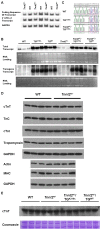
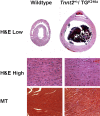
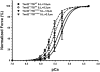
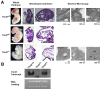
Methods and findings: We ablated Tnnt2 to produce heterozygous Tnnt2(+/-) mice, and crossbreeding produced homozygous null Tnnt2(-/-) embryos. We also generated transgenic mice overexpressing wildtype (TG(WT)) or DCM mutant (TG(K210Delta)) Tnnt2. Crossbreeding produced mice lacking one allele of Tnnt2, but carrying wildtype (Tnnt2(+/-)/TG(WT)) or mutant (Tnnt2(+/-)/TG(K210Delta)) transgenes. Tnnt2(+/-) mice relative to wildtype had significantly reduced transcript (0.82+/-0.06[SD] vs. 1.00+/-0.12 arbitrary units; p = 0.025), but not protein (1.01+/-0.20 vs. 1.00+/-0.13 arbitrary units; p = 0.44). Tnnt2(+/-) mice had normal hearts (histology, mass, left ventricular end diastolic diameter [LVEDD], fractional shortening [FS]). Moreover, whereas Tnnt2(+/-)/TG(K210Delta) mice had severe DCM, TG(K210Delta) mice had only mild DCM (FS 18+/-4 vs. 29+/-7%; p<0.01). The difference in severity of DCM may be attributable to a greater ratio of mutant to wildtype Tnnt2 transcript in Tnnt2(+/-)/TG(K210Delta) relative to TG(K210Delta) mice (2.42+/-0.08, p = 0.03). Tnnt2(+/-)/TG(K210Delta) muscle showed Ca(2+) desensitization (pCa(50) = 5.34+/-0.08 vs. 5.58+/-0.03 at sarcomere length 1.9 microm, p<0.01), but no difference in maximum force generation. Day 9.5 Tnnt2(-/-) embryos had normally looped hearts, but thin ventricular walls, large pericardial effusions, noncontractile hearts, and severely disorganized sarcomeres.
Conclusions: Absence of one Tnnt2 allele leads to a mild deficit in transcript but not protein, leading to a normal cardiac phenotype. DCM results from abnormal function of a mutant protein, which is associated with myocyte Ca(2+) desensitization. The severity of DCM depends on the ratio of mutant to wildtype Tnnt2 transcript. cTnT is essential for sarcomere formation, but normal embryonic heart looping occurs without contractile activity.
Conflict of interest statement
Figures

References
-
- Ahmad F, Seidman JG, Seidman CE. The genetic basis for cardiac remodeling. Annu Rev Genomics Hum Genet. 2005;6:185–216. - PubMed
-
- Kamisago M, Sharma SD, DePalma SR, Solomon S, Sharma P, et al. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. N Engl J Med. 2000;343:1688–96. - PubMed
-
- Blanchard EM, Iizuka K, Christe M, Conner DA, Geisterfer-Lowrance A, et al. Targeted ablation of the murine alpha-tropomyosin gene. Circ Res. 1997;81:1005–10. - PubMed
-
- Rethinasamy P, Muthuchamy M, Hewett T, Boivin G, Wolska BM, et al. Molecular and physiological effects of alpha-tropomyosin ablation in the mouse. Circ Res. 1998;82:116–23. - PubMed
-
- Harris SP, Bartley CR, Hacker TA, McDonald KS, Douglas PS, et al. Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ Res. 2002;90:594–601. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

