Structural basis for the autoinhibition of talin in regulating integrin activation
- PMID: 18614051
- PMCID: PMC2522368
- DOI: 10.1016/j.molcel.2008.06.011
Structural basis for the autoinhibition of talin in regulating integrin activation
Abstract
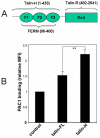
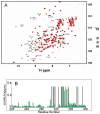
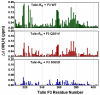
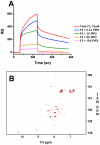
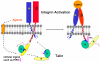
Activation of heterodimeric (alpha/beta) integrin transmembrane receptors by the 270 kDa cytoskeletal protein talin is essential for many important cell adhesive and physiological responses. A key step in this process involves interaction of phosphotyrosine-binding (PTB) domain in the N-terminal head of talin (talin-H) with integrin beta membrane-proximal cytoplasmic tails (beta-MP-CTs). Compared to talin-H, intact talin exhibits low potency in inducing integrin activation. Using NMR spectroscopy, we show that the large C-terminal rod domain of talin (talin-R) interacts with talin-H and allosterically restrains talin in a closed conformation. We further demonstrate that talin-R specifically masks a region in talin-PTB where integrin beta-MP-CT binds and competes with it for binding to talin-PTB. The inhibitory interaction is disrupted by a constitutively activating mutation (M319A) or by phosphatidylinositol 4,5-bisphosphate, a known talin activator. These data define a distinct autoinhibition mechanism for talin and suggest how it controls integrin activation and cell adhesion.
Figures


References
-
- Bax A, Grzesiek S. Methodological advances in protein NMR. Accounts Chem. Res. 1993;26:131–138.
-
- Bompard G, Martin M, Roy C, Vignon F, Freiss G. Membrane targeting of protein tyrosine phosphatase PTPL1 through its FERM domain via binding to phosphatidylinositol 4,5-biphosphate. J Cell Sci. 2003;116(Pt 12):2519–30. - PubMed
-
- Bonvin AM, Boelens R, Kaptein R. NMR analysis of protein interactions. Curr Opin Chem Biol. 2005;9(5):501–8. - PubMed
-
- Bouaouina M, Lad Y, Calderwood DA. The N-terminal domains of talin co-operate with the PTB-like domain to activate beta 1 and beta 3 integrins. J Biol Chem. 2007 Epub ahead of print. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

