Epstein-Barr virus BART microRNAs are produced from a large intron prior to splicing
- PMID: 18614630
- PMCID: PMC2546912
- DOI: 10.1128/JVI.00785-08
Epstein-Barr virus BART microRNAs are produced from a large intron prior to splicing
Abstract
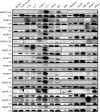
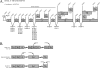
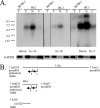
Latent Epstein-Barr virus (EBV) infection is associated with several lymphoproliferative disorders, including posttransplant lymphoma, Hodgkin's disease, and Burkitt's lymphoma, as well as nasopharyngeal carcinoma (NPC). Twenty-nine microRNAs (miRNAs) have been identified that are transcribed during latent infection from three clusters in the EBV genome. Two of the three clusters of miRNAs are made from the BamHI A rightward transcripts (BARTs), a set of alternatively spliced transcripts that are highly abundant in NPC but have not been shown to produce a detectable protein. This study indicates that while the BART miRNAs are located in the first four introns of the transcripts, processing of the pre-miRNAs from the primary transcript occurs prior to completion of the splicing reaction. Additionally, production of the BART miRNAs correlates with accumulation of a spliced mRNA in which exon 1 is joined directly to exon 3, suggesting that this form of the transcript may favor production of miRNAs. Sequence variations and processing of pre-miRNAs to the mature form also may account for various differences in miRNA abundance. Importantly, residual intronic pieces that result from processing of the pre-miRNAs were detected in the nucleus. The predicted structures of these pieces suggest there is a bias or temporal pattern to the production of the individual pre-miRNAs. These findings indicate that multiple factors contribute to the production of the BART miRNAs and to the apparent differences in abundance between the individual miRNAs of the cluster.
Figures
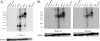




References
-
- Busson, P., G. Ganem, P. Flores, F. Mugneret, B. Clausse, B. Caillou, K. Braham, H. Wakasugi, M. Lipinski, and T. Tursz. 1988. Establishment and characterization of three transplantable EBV-containing nasopharyngeal carcinomas. Int. J. Cancer 42599-606. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

