Bacterial ApbC can bind and effectively transfer iron-sulfur clusters
- PMID: 18616280
- PMCID: PMC2562686
- DOI: 10.1021/bi800551y
Bacterial ApbC can bind and effectively transfer iron-sulfur clusters
Abstract
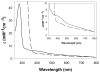
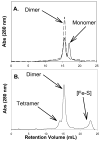
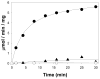
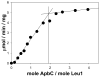
The metabolism of iron-sulfur ([Fe-S]) clusters requires a complex set of machinery that is still being defined. Mutants of Salmonella enterica lacking apbC have nutritional and biochemical properties indicative of defects in [Fe-S] cluster metabolism. ApbC is a 40.8 kDa homodimeric ATPase and as purified contains little iron and no acid-labile sulfide. An [Fe-S] cluster was reconstituted on ApbC, generating a protein that bound 2 mol of Fe and 2 mol of S (2-) per ApbC monomer and had a UV-visible absorption spectrum similar to known [4Fe-4S] cluster proteins. Holo-ApbC could rapidly and effectively activate Saccharomyces cerevisiae apo-isopropylmalate isolomerase (Leu1) in vitro, a process known to require the transfer of a [4Fe-4S] cluster. Maximum activation was achieved with 2 mol of ApbC per 1 mol of apo-Leu1. This article describes the first biochemical activity of ApbC in the context of [Fe-S] cluster metabolism. The data herein support a model in which ApbC coordinates an [4Fe-4S] cluster across its dimer interface and can transfer this cluster to an apoprotein acting as an [Fe-S] cluster scaffold protein, a function recently deduced for its eukaryotic homologues.
Figures

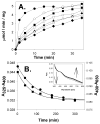
References
-
- Lill R, Muhlenhoff U. Iron-sulfur protein biogenesis in eukaryotes: components and mechanisms. Annu Rev Cell Dev Biol. 2006;22:457–486. - PubMed
-
- Beinert H. Iron-sulfur proteins: ancient structures, still full of surprises. Journal of Biological and Inorganic Chemistry. 2000;5:2–15. - PubMed
-
- Frazzon J, Dean DR. Formation of iron-sulfur clusters in bacteria: an emerging field in bioinorganic chemistry. Current Opinion in Chemical Biology. 2003;7:166–173. - PubMed
-
- Takahashi Y, Tokumoto U. A third bacterial system for the assembly of iron-sulfur clusters with homologs in archaea and plastids. J Biol Chem. 2002;277:28380–28393. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

