The loci of evolution: how predictable is genetic evolution?
- PMID: 18616572
- PMCID: PMC2613234
- DOI: 10.1111/j.1558-5646.2008.00450.x
The loci of evolution: how predictable is genetic evolution?
Abstract
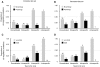
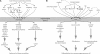
Is genetic evolution predictable? Evolutionary developmental biologists have argued that, at least for morphological traits, the answer is a resounding yes. Most mutations causing morphological variation are expected to reside in the cis-regulatory, rather than the coding, regions of developmental genes. This "cis-regulatory hypothesis" has recently come under attack. In this review, we first describe and critique the arguments that have been proposed in support of the cis-regulatory hypothesis. We then test the empirical support for the cis-regulatory hypothesis with a comprehensive survey of mutations responsible for phenotypic evolution in multicellular organisms. Cis-regulatory mutations currently represent approximately 22% of 331 identified genetic changes although the number of cis-regulatory changes published annually is rapidly increasing. Above the species level, cis-regulatory mutations altering morphology are more common than coding changes. Also, above the species level cis-regulatory mutations predominate for genes not involved in terminal differentiation. These patterns imply that the simple question "Do coding or cis-regulatory mutations cause more phenotypic evolution?" hides more interesting phenomena. Evolution in different kinds of populations and over different durations may result in selection of different kinds of mutations. Predicting the genetic basis of evolution requires a comprehensive synthesis of molecular developmental biology and population genetics.
Figures


References
-
- Abzhanov A, Protas M, Grant B R, Grant P R, Tabin C J. Bmp4 and morphological variation of beaks in Darwin's finches. Science. 2004;305:1462–1465. - PubMed
-
- Adamowicz S J, Purvis A. From more to fewer? Testing an allegedly pervasive trend in the evolution of morphological structure. Evol. Int. J. Org. Evol. 2006;60:1402–1416. - PubMed
-
- Akam M. Hox genes and the evolution of diverse body plans. Philos. Trans. R. Society Lond. B. 1995;349:313–319. - PubMed
-
- Akam M. Hox genes, homeosis and the evolution of segment identity: no need for hopeless monsters. Int. J. Develop. Biol. 1998;42:445–451. - PubMed
-
- Alonso C R, Wilkins A S. The molecular elements that underlie developmental evolution. Nat. Rev. Genet. 2005;6:709–715. - PubMed
