MALDI imaging mass spectrometry for direct tissue analysis: a new frontier for molecular histology
- PMID: 18618129
- PMCID: PMC2522327
- DOI: 10.1007/s00418-008-0469-9
MALDI imaging mass spectrometry for direct tissue analysis: a new frontier for molecular histology
Abstract
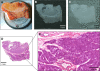
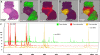
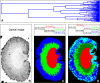
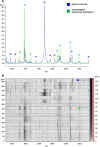
Matrix-assisted laser desorption/ionization (MALDI) imaging mass spectrometry (IMS) is a powerful tool for investigating the distribution of proteins and small molecules within biological systems through the in situ analysis of tissue sections. MALDI-IMS can determine the distribution of hundreds of unknown compounds in a single measurement and enables the acquisition of cellular expression profiles while maintaining the cellular and molecular integrity. In recent years, a great many advances in the practice of imaging mass spectrometry have taken place, making the technique more sensitive, robust, and ultimately useful. In this review, we focus on the current state of the art of MALDI-IMS, describe basic technological developments for MALDI-IMS of animal and human tissues, and discuss some recent applications in basic research and in clinical settings.
Figures


References
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1021/cr990076h', 'is_inner': False, 'url': 'https://doi.org/10.1021/cr990076h'}, {'type': 'PubMed', 'value': '11712248', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/11712248/'}]}
- Aebersold R, Goodlett DR (2001) Mass spectrometry in proteomics. Chem Rev 101(2):269–295 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1021/ac051534r', 'is_inner': False, 'url': 'https://doi.org/10.1021/ac051534r'}, {'type': 'PubMed', 'value': '16448057', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/16448057/'}]}
- Aerni HR, Cornett DS, Caprioli RM (2006) Automated acoustic matrix deposition for MALDI sample preparation. Anal Chem 78(3):827–834 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1021/ac048329g', 'is_inner': False, 'url': 'https://doi.org/10.1021/ac048329g'}, {'type': 'PubMed', 'value': '15679338', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/15679338/'}]}
- Altelaar AF, van Minnen J, Jiménez CR, Heeren RM, Piersma SR (2005) Direct molecular imaging of Lymnaea stagnalis nervous tissue at subcellular spatial resolution by mass spectrometry. Anal Chem 77(3):735–741 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1021/ac0513111', 'is_inner': False, 'url': 'https://doi.org/10.1021/ac0513111'}, {'type': 'PubMed', 'value': '16448046', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/16448046/'}]}
- Altelaar AF, Klinkert I, Jalink K, de Lange RP, Adan RA, Heeren RM, Piersma SR (2006) Gold-enhanced biomolecular surface imaging of cells and tissue by SIMS and MALDI mass spectrometry. Anal Chem 78(3):734–742 - PubMed
-
- {'text': '', 'ref_index': 1, 'ids': [{'type': 'DOI', 'value': '10.1038/nprot.2007.117', 'is_inner': False, 'url': 'https://doi.org/10.1038/nprot.2007.117'}, {'type': 'PubMed', 'value': '17546014', 'is_inner': True, 'url': 'https://pubmed.ncbi.nlm.nih.gov/17546014/'}]}
- Altelaar AF, Luxembourg SL, McDonnell LA, Piersma SR, Heeren RM (2007) Imaging mass spectrometry at cellular length scales. Nat Protoc 2(5):1185–1196 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

