Duffy antigen receptor for chemokines mediates trans-infection of HIV-1 from red blood cells to target cells and affects HIV-AIDS susceptibility
- PMID: 18621010
- PMCID: PMC2562426
- DOI: 10.1016/j.chom.2008.06.002
Duffy antigen receptor for chemokines mediates trans-infection of HIV-1 from red blood cells to target cells and affects HIV-AIDS susceptibility
Abstract
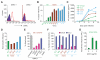
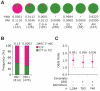
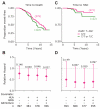
Duffy antigen receptor for chemokines (DARC) expressed on red blood cells (RBCs) influences plasma levels of HIV-1-suppressive and proinflammatory chemokines such as CCL5/RANTES. DARC is also the RBC receptor for Plasmodium vivax. Africans with DARC -46C/C genotype, which confers a DARC-negative phenotype, are resistant to vivax malaria. Here, we show that HIV-1 attaches to RBCs via DARC, effecting trans-infection of target cells. In African Americans, DARC -46C/C is associated with 40% increase in the odds of acquiring HIV-1. If extrapolated to Africans, approximately 11% of the HIV-1 burden in Africa may be linked to this genotype. After infection occurs, however, DARC-negative RBC status is associated with slower disease progression. Furthermore, the disease-accelerating effect of a previously described CCL5 polymorphism is evident only in DARC-expressing and not in DARC-negative HIV-infected individuals. Thus, DARC influences HIV/AIDS susceptibility by mediating trans-infection of HIV-1 and by affecting both chemokine-HIV interactions and chemokine-driven inflammation.
Figures
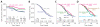

Comment in
-
HIV and chemokine binding to red blood cells--DARC matters.Cell Host Microbe. 2008 Jul 17;4(1):3-5. doi: 10.1016/j.chom.2008.06.006. Cell Host Microbe. 2008. PMID: 18621004 Review.
-
The Duffy antigen receptor for chemokines null promoter variant does not influence HIV-1 acquisition or disease progression.Cell Host Microbe. 2009 May 8;5(5):408-10; author reply 418-9. doi: 10.1016/j.chom.2009.04.011. Cell Host Microbe. 2009. PMID: 19454339 Free PMC article.
-
Expression of Duffy antigen receptor for chemokines (DARC) has no effect on HIV-1 acquisition or progression to AIDS in African Americans.Cell Host Microbe. 2009 May 8;5(5):411-3; author reply 418-9. doi: 10.1016/j.chom.2009.04.010. Cell Host Microbe. 2009. PMID: 19454340 Free PMC article.
-
Lack of Duffy antigen receptor for chemokines: no influence on HIV disease progression in an African treatment-naive population.Cell Host Microbe. 2009 May 8;5(5):413-5; author reply 418-9. doi: 10.1016/j.chom.2009.04.009. Cell Host Microbe. 2009. PMID: 19454341 Free PMC article. No abstract available.
-
Duffy antigen polymorphisms do not alter progression of HIV in African Americans in the MACS cohort.Cell Host Microbe. 2009 May 8;5(5):415-7; author reply 418-9. doi: 10.1016/j.chom.2009.04.013. Cell Host Microbe. 2009. PMID: 19454342 No abstract available.
References
-
- Abu-Raddad LJ, Patnaik P, Kublin JG. Dual infection with HIV and malaria fuels the spread of both diseases in sub-Saharan Africa. Science. 2006;314:1603–1606. - PubMed
-
- Banki Z, Wilflingseder D, Ammann CG, Pruenster M, Mullauer B, Hollander K, Meyer M, Sprinzl GM, van Lunzen J, Stellbrink HJ, et al. Factor I-mediated processing of complement fragments on HIV immune complexes targets HIV to CR2-expressing B cells and facilitates B cell-mediated transmission of opsonized HIV to T cells. J Immunol. 2006;177:3469–3476. - PubMed
-
- Buve A. HIV epidemics in Africa: what explains the variations in HIV prevalence? IUBMB Life. 2002;53:193–195. - PubMed
-
- Comerford I, Nibbs RJ. Post-translational control of chemokines: a role for decoy receptors? Immunol Lett. 2005;96:163–174. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

