Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome
- PMID: 18621012
- PMCID: PMC3519368
- DOI: 10.1016/j.chom.2008.05.013
Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome
Abstract
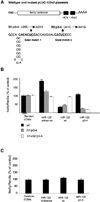
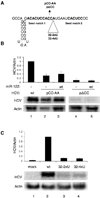
MicroRNAs usually interact with 3' noncoding regions (3'NCRs) of target mRNAs leading to downregulation of mRNA expression. In contrast, liver-specific microRNA miR-122 interacts with the 5' end of the hepatitis C virus RNA genome, resulting in increased viral RNA abundance. We find that inserting the viral miR-122 binding site into the 3' noncoding region of a reporter mRNA leads to downregulation of mRNA expression, indicating that the location of the miR-122 binding site dictates its effect on gene regulation. Furthermore, we discovered an adjacent, second miR-122 binding site, separated from the first by a highly conserved 14-nucleotide sequence. Mutational analysis demonstrates that both miR-122 binding sites in a single viral genome are occupied by the microRNA and function cooperatively to regulate target gene expression. These findings set a paradigm for dual, position-dependent functions of tandem microRNA-binding sites. Targeting an oligomeric microRNA complex offers potential as an antiviral-intervention strategy.
Figures





References
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Smith JA, Seidman JG, Struhl K. Current Protocols in Molecular Biology. New York: Current Protocols (Greene Publishing Associates, Inc. and John Wiley & Sons, Inc.; 1989.
-
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. - PubMed
-
- Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biology. 2004;1:106–113. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

