Identifying autism loci and genes by tracing recent shared ancestry
- PMID: 18621663
- PMCID: PMC2586171
- DOI: 10.1126/science.1157657
Identifying autism loci and genes by tracing recent shared ancestry
Erratum in
- Science. 2010 Dec 24;330(6012):1746
Abstract
To find inherited causes of autism-spectrum disorders, we studied families in which parents share ancestors, enhancing the role of inherited factors. We mapped several loci, some containing large, inherited, homozygous deletions that are likely mutations. The largest deletions implicated genes, including PCDH10 (protocadherin 10) and DIA1 (deleted in autism1, or c3orf58), whose level of expression changes in response to neuronal activity, a marker of genes involved in synaptic changes that underlie learning. A subset of genes, including NHE9 (Na+/H+ exchanger 9), showed additional potential mutations in patients with unrelated parents. Our findings highlight the utility of "homozygosity mapping" in heterogeneous disorders like autism but also suggest that defective regulation of gene expression after neural activity may be a mechanism common to seemingly diverse autism mutations.
Figures

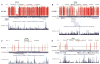

Comment in
-
Genetics. Insights into the pathogenesis of autism.Science. 2008 Jul 11;321(5886):208-9. doi: 10.1126/science.1160555. Science. 2008. PMID: 18621658 No abstract available.
-
Autistic phenotype from MEF2C knockout cells.Science. 2009 Jan 9;323(5911):208. doi: 10.1126/science.323.5911.208b. Science. 2009. PMID: 19131610 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
- MH64547/MH/NIMH NIH HHS/United States
- R01 MH083565/MH/NIMH NIH HHS/United States
- 5P30HD018655-26/HD/NICHD NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- 1K01MH71801/MH/NIMH NIH HHS/United States
- 1R01 MH083565/MH/NIMH NIH HHS/United States
- K23 MH080954/MH/NIMH NIH HHS/United States
- 1K23MH080954-01/MH/NIMH NIH HHS/United States
- 5R01NS048276-05/NS/NINDS NIH HHS/United States
- R01 NS048276/NS/NINDS NIH HHS/United States
- K01 MH071801/MH/NIMH NIH HHS/United States
- N01-HG-65403/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

