Genetic determinants of self identity and social recognition in bacteria
- PMID: 18621670
- PMCID: PMC2567286
- DOI: 10.1126/science.1160033
Genetic determinants of self identity and social recognition in bacteria
Abstract
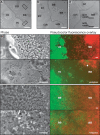
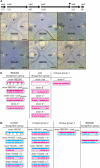
The bacterium Proteus mirabilis is capable of movement on solid surfaces by a type of motility called swarming. Boundaries form between swarming colonies of different P. mirabilis strains but not between colonies of a single strain. A fundamental requirement for boundary formation is the ability to discriminate between self and nonself. We have isolated mutants that form boundaries with their parent. The mutations map within a six-gene locus that we term ids for identification of self. Five of the genes in the ids locus are required for recognition of the parent strain as self. Three of the ids genes are interchangeable between strains, and two encode specific molecular identifiers.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

