Coordinated evolution of the hepatitis C virus
- PMID: 18621679
- PMCID: PMC2474538
- DOI: 10.1073/pnas.0801774105
Coordinated evolution of the hepatitis C virus
Abstract
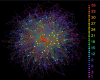
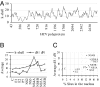
Hepatitis C virus is a genetically heterogeneous RNA virus that is a major cause of liver disease worldwide. Here, we show that, despite its extensive heterogeneity, the evolution of hepatitis C virus is primarily shaped by negative selection and that numerous coordinated substitutions in the polyprotein can be organized into a scale-free network whose degree of connections between sites follows a power-law distribution. This network shares all major properties with many complex biological and technological networks. The topological structure and hierarchical organization of this network suggest that a small number of amino acid sites exert extensive impact on hepatitis C virus evolution. Nonstructural proteins are enriched for negatively selected sites of high centrality, whereas structural proteins are enriched for positively selected sites located in the periphery of the network. The complex network of coordinated substitutions is an emergent property of genetic systems with implications for evolution, vaccine research, and drug development. In addition to such properties as polymorphism or strength of selection, the epistatic connectivity mapped in the network is important for typing individual sites, proteins, or entire genetic systems. The network topology may help devise molecular intervention strategies for disrupting viral functions or impeding compensatory changes for vaccine escape or drug resistance mutations. Also, it may be used to find new therapeutic targets, as suggested in this study for the NS4A protein, which plays an important role in the network.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Alberti A, Chemello L, Benvegnu L. Natural history of hepatitis C. J Hepatol. 1999;31:17–24. - PubMed
-
- Bowen D, Walker C. Adaptive immune responses in acute and chronic hepatitis C virus infection. Nature. 2005;436:946–952. - PubMed
-
- Choo Q, et al. Isolation Of A Cdna clone derived from a bloodborne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. - PubMed
-
- Roingeard P, Hourioux C, Blanchard E, Brand D, Ait-Goughoulte M. Hepatitis C virus ultrastructure and morphogenesis. Biol Cell. 2004;96:103–108. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

