Crystal structures of substrate-bound and substrate-free cytochrome P450 46A1, the principal cholesterol hydroxylase in the brain
- PMID: 18621681
- PMCID: PMC2474539
- DOI: 10.1073/pnas.0803717105
Crystal structures of substrate-bound and substrate-free cytochrome P450 46A1, the principal cholesterol hydroxylase in the brain
Abstract
By converting cholesterol to 24S-hydroxycholesterol, cytochrome P450 46A1 (CYP46A1) initiates the major pathway for cholesterol removal from the brain. Two crystal structures of CYP46A1 were determined. First is the 1.9-A structure of CYP46A1 complexed with a high-affinity substrate cholesterol 3-sulfate (CH-3S). The second structure is that of the substrate-free CYP46A1 at 2.4-A resolution. CH-3S is bound in the productive orientation and occupies the entire length of the banana-shaped hydrophobic active-site cavity. A unique helix B'-C loop insertion (residues 116-120) contributes to positioning cholesterol for oxygenation catalyzed by CYP46A1. A comparison with the substrate-free structure reveals substantial substrate-induced conformational changes in CYP46A1 and suggests that structurally distinct compounds could bind in the enzyme active site. In vitro assays were performed to characterize the effect of different therapeutic agents on cholesterol hydroxylase activity of purified full-length recombinant CYP46A1, and several strong inhibitors and modest coactivators of CYP46A1 were identified. Structural and biochemical data provide evidence that CYP46A1 activity could be altered by exposure to some therapeutic drugs and potentially other xenobiotics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
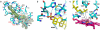
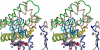
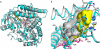
References
-
- Dietschy JM, Turley SD. Cholesterol metabolism in the brain. Curr Opin Lipidol. 2001;12:105–112. - PubMed
-
- Bjorkhem I, Meaney S. Brain cholesterol: Long secret life behind a barrier. Arterioscler Thromb Vasc Biol. 2004;24:806–815. - PubMed
-
- Wolozin B. Cholesterol and the biology of Alzheimer's disease. Neuron. 2004;41:7–10. - PubMed
-
- Carter CJ. Convergence of genes implicated in Alzheimer's disease on the cerebral cholesterol shuttle: APP, cholesterol, lipoproteins, and atherosclerosis. Neurochem Int. 2007;50:12–38. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

