Alpha-Helix folding in the presence of structural constraints
- PMID: 18621686
- PMCID: PMC2474473
- DOI: 10.1073/pnas.0712099105
Alpha-Helix folding in the presence of structural constraints
Abstract
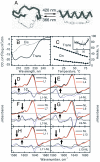
We have investigated the site-specific folding kinetics of a photoswitchable cross-linked alpha-helical peptide by using single (13)C = (18)O isotope labeling together with time-resolved IR spectroscopy. We observe that the folding times differ from site to site by a factor of eight at low temperatures (6 degrees C), whereas at high temperatures (45 degrees C), the spread is considerably smaller. The trivial sum of the site signals coincides with the overall folding signal of the unlabeled peptide, and different sites fold in a noncooperative manner. Moreover, one of the sites exhibits a decrease of hydrogen bonding upon folding, implying that the unfolded state at low temperature is not unstructured. Molecular dynamics simulations at low temperature reveal a stretched-exponential behavior which originates from parallel folding routes that start from a kinetically partitioned unfolded ensemble. Different metastable structures (i.e., traps) in the unfolded ensemble have a different ratio of loop and helical content. Control simulations of the peptide at high temperature, as well as without the cross-linker at low temperature, show faster and simpler (i.e., single-exponential) folding kinetics. The experimental and simulation results together provide strong evidence that the rate-limiting step in formation of a structurally constrained alpha-helix is the escape from heterogeneous traps rather than the nucleation rate. This conclusion has important implications for an alpha-helical segment within a protein, rather than an isolated alpha-helix, because the cross-linker is a structural constraint similar to those present during the folding of a globular protein.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Eaton WA, Henry ER, Hofrichter J, Mozzarelli A. Is cooperative oxygen binding by hemoglobin really understood? Nat Struct Biol. 1999;6:351–358. - PubMed
-
- Creighton TE. Proteins. New York: Freeman; 1993.
-
- Zimm BH, Bragg JK. Theory of the phase transition between helix and random coil in polypeptide chains. J Chem Phys. 1959;31:526–535.
-
- Lifson S, Roig A. On the theory of helix–coil transition in polypeptides. J Chem Phys. 1961;34:1963–1974.
-
- Scholtz JM, Qian H, York EJ, Stewart JM, Baldwin RL. Parameters of helix–coil transition theory for alanine-based peptides of varying chain lengths in water. Biopolymers. 1991;31:1463–1470. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

