Experience using web services for biological sequence analysis
- PMID: 18621748
- PMCID: PMC2989672
- DOI: 10.1093/bib/bbn029
Experience using web services for biological sequence analysis
Abstract
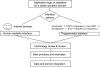
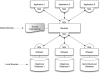
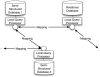
Programmatic access to data and tools through the web using so-called web services has an important role to play in bioinformatics. In this article, we discuss the most popular approaches based on SOAP/WS-I and REST and describe our, a cross section of the community, experiences with providing and using web services in the context of biological sequence analysis. We briefly review main technological approaches as well as best practice hints that are useful for both users and developers. Finally, syntactic and semantic data integration issues with multiple web services are discussed.
Figures

References
-
- WC3 Web Services. [(January 2008, date last accessed).]. http://www.w3.org/2002/ws/
-
- EMBRACE Consortium. Workshop: Deploying Web Services for Biological Sequence Annotation (Geneva, Switzerland, from 30 May to 1 June 2007) [(December 2007, date last accessed).]. Deliverable D5.2.10, June 2007. Available at http://www.embracegrid.info.
-
- EMBRACE. Network of Excellence. [(October 2007, date last accessed).]. http://www.embracegrid.info.
-
- Gattiker A, Michoud K, Rivoire C, et al. Automated annotation of microbial proteomes in Swiss-Prot. Comput Biol Chem. 2003;27(1):49–58. - PubMed

