Effects of multiple-bond ruptures on kinetic parameters extracted from force spectroscopy measurements: revisiting biotin-streptavidin interactions
- PMID: 18621812
- PMCID: PMC2553120
- DOI: 10.1529/biophysj.108.133900
Effects of multiple-bond ruptures on kinetic parameters extracted from force spectroscopy measurements: revisiting biotin-streptavidin interactions
Abstract
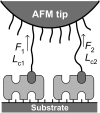
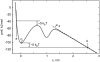
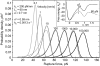
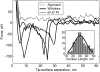
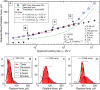
Force spectroscopy measurements of the rupture of the molecular bond between biotin and streptavidin often results in a wide distribution of rupture forces. We attribute the long tail of high rupture forces to the nearly simultaneous rupture of more than one molecular bond. To decrease the number of possible bonds, we employed hydrophilic polymeric tethers to attach biotin molecules to the atomic force microscope probe. It is shown that the measured distributions of rupture forces still contain high forces that cannot be described by the forced dissociation from a deep potential well. We employed a recently developed analytical model of simultaneous rupture of two bonds connected by polymer tethers with uneven length to fit the measured distributions. The resulting kinetic parameters agree with the energy landscape predicted by molecular dynamics simulations. It is demonstrated that when more than one molecular bond might rupture during the pulling measurements there is a noise-limited range of probe velocities where the kinetic parameters measured by force spectroscopy correspond to the true energy landscape. Outside this range of velocities, the kinetic parameters extracted by using the standard most probable force approach might be interpreted as artificial energy barriers that are not present in the actual energy landscape. Factors that affect the range of useful velocities are discussed.
Figures

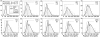
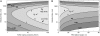
Similar articles
-
Association kinetics from single molecule force spectroscopy measurements.Biophys J. 2009 Apr 22;96(8):3412-22. doi: 10.1016/j.bpj.2009.01.031. Biophys J. 2009. PMID: 19383484 Free PMC article.
-
Energy landscape of streptavidin-biotin complexes measured by atomic force microscopy.Biochemistry. 2000 Aug 22;39(33):10219-23. doi: 10.1021/bi992715o. Biochemistry. 2000. PMID: 10956011
-
Energy landscape roughness of the streptavidin-biotin interaction.J Mol Recognit. 2007 Nov-Dec;20(6):495-501. doi: 10.1002/jmr.841. J Mol Recognit. 2007. PMID: 17902095
-
Probing the relation between force--lifetime--and chemistry in single molecular bonds.Annu Rev Biophys Biomol Struct. 2001;30:105-28. doi: 10.1146/annurev.biophys.30.1.105. Annu Rev Biophys Biomol Struct. 2001. PMID: 11340054 Review.
-
Practical single molecule force spectroscopy: how to determine fundamental thermodynamic parameters of intermolecular bonds with an atomic force microscope.Methods. 2013 Apr 1;60(2):142-50. doi: 10.1016/j.ymeth.2013.03.014. Epub 2013 Mar 24. Methods. 2013. PMID: 23531626 Review.
Cited by
-
Adhesion of mussel foot protein Mefp-5 to mica: an underwater superglue.Biochemistry. 2012 Aug 21;51(33):6511-8. doi: 10.1021/bi3002538. Epub 2012 Aug 8. Biochemistry. 2012. PMID: 22873939 Free PMC article.
-
Direct Reading of Bona Fide Barcode Assays for Diagnostics with Smartphone Apps.Sci Rep. 2015 Jun 30;5:11727. doi: 10.1038/srep11727. Sci Rep. 2015. PMID: 26122608 Free PMC article.
-
Hidden multiple bond effects in dynamic force spectroscopy.Biophys J. 2012 Mar 7;102(5):1184-93. doi: 10.1016/j.bpj.2012.01.037. Epub 2012 Mar 6. Biophys J. 2012. PMID: 22404941 Free PMC article.
-
Rupture of multiple receptor-ligand bonds: bimodal distribution of bond rupture force.Eur Phys J E Soft Matter. 2012 Sep;35(9):94. doi: 10.1140/epje/i2012-12094-9. Epub 2012 Sep 27. Eur Phys J E Soft Matter. 2012. PMID: 23015276
-
Kinetics of the multistep rupture of fibrin 'A-a' polymerization interactions measured using atomic force microscopy.Biophys J. 2009 Nov 18;97(10):2820-8. doi: 10.1016/j.bpj.2009.08.042. Biophys J. 2009. PMID: 19917237 Free PMC article.
References
-
- Bell, G. I. 1978. Models for specific adhesion of cells to cells. Science. 200:618–627. - PubMed
-
- Baltz, J. M., and R. A. Cone. 1990. The strength of non-covalent biological bonds and adhesions by multiple independent bonds. J. Theor. Biol. 142:163–178. - PubMed
-
- Leckband, D., and J. Israelachvili. 2001. Intermolecular forces in biology. Q. Rev. Biophys. 34:105–267. - PubMed
-
- Garg, A. 1995. Escape-field distribution for escape from a metastable potential well subject to a steadily increasing bias field. Phys. Rev. B. 51:15592–15595. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

