Metallic nanoparticles used to estimate the structural integrity of DNA motifs
- PMID: 18621817
- PMCID: PMC2547441
- DOI: 10.1529/biophysj.108.138479
Metallic nanoparticles used to estimate the structural integrity of DNA motifs
Abstract
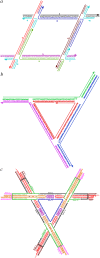
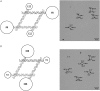
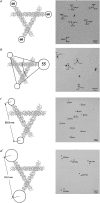
Branched DNA motifs can be designed to assume a variety of shapes and structures. These structures can be characterized by numerous solution techniques; the structures also can be inferred from atomic force microscopy of two-dimensional periodic arrays that the motifs form via cohesive interactions. Examples of these motifs are the DNA parallelogram, the bulged-junction DNA triangle, and the three-dimensional-double crossover (3D-DX) DNA triangle. The ability of these motifs to withstand stresses without changing geometrical structure is clearly of interest if the motif is to be used in nanomechanical devices or to organize other large chemical species. Metallic nanoparticles can be attached to DNA motifs, and the arrangement of these particles can be established by transmission electron microscopy. We have attached 5 nm or 10 nm gold nanoparticles to every vertex of DNA parallelograms, to two or three vertices of 3D-DX DNA triangle motifs, and to every vertex of bulged-junction DNA triangles. We demonstrate by transmission electron microscopy that the DNA parallelogram motif and the bulged-junction DNA triangle are deformed by the presence of the gold nanoparticles, whereas the structure of the 3D-DX DNA triangle motif appears to be minimally distorted. This method provides a way to estimate the robustness and potential utility of the many new DNA motifs that are becoming available.
Figures



References
-
- Chen, J., and N. C. Seeman. 1991. The synthesis from DNA of a molecule with the connectivity of a cube. Nature. 350:631–633. - PubMed
-
- Winfree, E., F. Liu, L. A. Wenzler, and N. C. Seeman. 1998. Design and self-assembly of two-dimensional DNA crystals. Nature. 394:539–544. - PubMed
-
- Mao, C., W. Sun, Z. Shen, and N. C. Seeman. 1999. A DNA nanomechanical device based on the B-Z transition. Nature. 397:144–146. - PubMed
-
- Yan, H., X. Zhang, Z. Shen, and N. C. Seeman. 2002. A robust DNA mechanical device controlled by hybridization topology. Nature. 415:62–65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

