A novel RNA-binding protein associated with cell plate formation
- PMID: 18621982
- PMCID: PMC2528124
- DOI: 10.1104/pp.108.120527
A novel RNA-binding protein associated with cell plate formation
Abstract
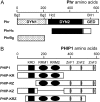
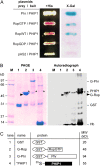
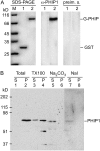
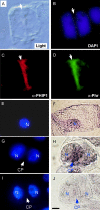
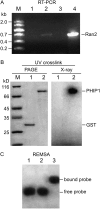
Building a cell plate during cytokinesis in plant cells requires the participation of a number of proteins in a multistep process. We previously identified phragmoplastin as a cell plate-specific protein involved in creating a tubulovesicular network at the cell plate. We report here the identification and characterization of a phragmoplastin-interacting protein, PHIP1, in Arabidopsis (Arabidopsis thaliana). It contains multiple functional motifs, including a lysine-rich domain, two RNA recognition motifs, and three CCHC-type zinc fingers. Polypeptides with similar motif structures were found only in plant protein databases, but not in the sequenced prokaryotic, fungal, and animal genomes, suggesting that PHIP1 represents a plant-specific RNA-binding protein. In addition to phragmoplastin, two Arabidopsis small GTP-binding proteins, Rop1 and Ran2, are also found to interact with PHIP1. The zinc fingers of PHIP1 were not required for its interaction with Rop1 and phragmoplastin, but they may participate in its binding with the Ran2 mRNA. Immunofluorescence, in situ RNA hybridization, and green fluorescent protein tagging experiments showed the association of PHIP1 with the forming cell plate during cytokinesis. Taken together, our data suggest that PHIP1 is a novel RNA-binding protein and may play a unique role in the polarized mRNA transport to the vicinity of the cell plate.
Figures

Similar articles
-
Arabidopsis dynamin-related protein DRP2B is co-localized with DRP1A on the leading edge of the forming cell plate.Plant Cell Rep. 2008 Oct;27(10):1581-6. doi: 10.1007/s00299-008-0583-0. Epub 2008 Jul 9. Plant Cell Rep. 2008. PMID: 18612642
-
Phragmoplastin dynamics: multiple forms, microtubule association and their roles in cell plate formation in plants.Plant Mol Biol. 2003 Oct;53(3):297-312. doi: 10.1023/b:plan.0000006936.50532.3a. Plant Mol Biol. 2003. PMID: 14750520
-
A novel UDP-glucose transferase is part of the callose synthase complex and interacts with phragmoplastin at the forming cell plate.Plant Cell. 2001 Apr;13(4):769-79. doi: 10.1105/tpc.13.4.769. Plant Cell. 2001. PMID: 11283335 Free PMC article.
-
The role of ADP-ribosylation factor and SAR1 in vesicular trafficking in plants.Biochim Biophys Acta. 2004 Jul 1;1664(1):9-30. doi: 10.1016/j.bbamem.2004.04.005. Biochim Biophys Acta. 2004. PMID: 15238254 Review.
-
Pumilio Puf domain RNA-binding proteins in Arabidopsis.Plant Signal Behav. 2011 Mar;6(3):364-8. doi: 10.4161/psb.6.3.14380. Epub 2011 Mar 1. Plant Signal Behav. 2011. PMID: 21350339 Free PMC article. Review.
Cited by
-
The Small G Protein AtRAN1 Regulates Vegetative Growth and Stress Tolerance in Arabidopsis thaliana.PLoS One. 2016 Jun 3;11(6):e0154787. doi: 10.1371/journal.pone.0154787. eCollection 2016. PLoS One. 2016. PMID: 27258048 Free PMC article.
-
An ancient P-loop GTPase in rice is regulated by a higher plant-specific regulatory protein.J Biol Chem. 2010 Nov 26;285(48):37359-69. doi: 10.1074/jbc.M110.172080. Epub 2010 Sep 28. J Biol Chem. 2010. PMID: 20876569 Free PMC article.
-
Genome-wide analysis of CCHC-type zinc finger (ZCCHC) proteins in yeast, Arabidopsis, and humans.Cell Mol Life Sci. 2020 Oct;77(20):3991-4014. doi: 10.1007/s00018-020-03518-7. Epub 2020 Apr 18. Cell Mol Life Sci. 2020. PMID: 32303790 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

