Combination of novel green fluorescent protein mutant TSapphire and DsRed variant mOrange to set up a versatile in planta FRET-FLIM assay
- PMID: 18621983
- PMCID: PMC2528103
- DOI: 10.1104/pp.108.117358
Combination of novel green fluorescent protein mutant TSapphire and DsRed variant mOrange to set up a versatile in planta FRET-FLIM assay
Abstract
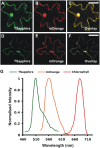
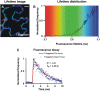
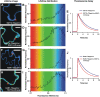
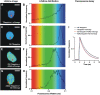
Förster resonance energy transfer (FRET) measurements based on fluorescence lifetime imaging microscopy (FLIM) are increasingly being used to assess molecular conformations and associations in living systems. Reduction in the excited-state lifetime of the donor fluorophore in the presence of an appropriately positioned acceptor is taken as strong evidence of FRET. Traditionally, cyan fluorescent protein has been widely used as a donor fluorophore in FRET experiments. However, given its photolabile nature, low quantum yield, and multiexponential lifetime, cyan fluorescent protein is far from an ideal donor in FRET imaging. Here, we report the application and use of the TSapphire mutant of green fluorescent protein as an efficient donor to mOrange in FLIM-based FRET imaging in intact plant cells. Using time-correlated single photon counting-FLIM, we show that TSapphire expressed in living plant cells decays with lifetime of 2.93 +/- 0.09 ns. Chimerically linked TSapphire and mOrange (with 16-amino acid linker in between) exhibit substantial energy transfer based on the reduction in the lifetime of TSapphire in the presence of the acceptor mOrange. Experiments performed with various genetically and/or biochemically known interacting plant proteins demonstrate the versatility of the FRET-FLIM system presented here in different subcellular compartments tested (cytosol, nucleus, and at plasma membrane). The better spectral overlap with red monomers, higher photostability, and monoexponential lifetime of TSapphire makes it an ideal FRET-FLIM donor to study protein-protein interactions in diverse eukaryotic systems overcoming, in particular, many technical challenges encountered (like autofluorescence of cell walls and fluorescence of pigments associated with photosynthetic apparatus) while studying plant protein dynamics and interactions.
Figures

References
-
- Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P (2002) The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295 2073–2076 - PubMed
-
- Bastiaens PI, Squire A (1999) Fluorescence lifetime imaging microscopy: spatial resolution of biochemical processes in the cell. Trends Cell Biol 9 48–52 - PubMed
-
- Becker W, Bergmann A, Haustein E, Petrasek Z, Schwille P, Biskup C, Kelbauskas L, Benndorf K, Klocker N, Anhut T, et al (2006) Fluorescence lifetime images and correlation spectra obtained by multidimensional time-correlated single photon counting. Microsc Res Tech 69 186–195 - PubMed
-
- Becker W, Bergmann A, Hink MA, Konig K, Benndorf K, Biskup C (2004) Fluorescence lifetime imaging by time-correlated single-photon counting. Microsc Res Tech 63 58–66 - PubMed
-
- Bhat RA, Borst JW, Riehl M, Thompson RD (2004) Interaction of maize Opaque-2 and the transcriptional co-activators GCN5 and ADA2, in the modulation of transcriptional activity. Plant Mol Biol 55 239–252 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

