Strategies for carbohydrate recognition by the mannose 6-phosphate receptors
- PMID: 18621992
- PMCID: PMC2733771
- DOI: 10.1093/glycob/cwn061
Strategies for carbohydrate recognition by the mannose 6-phosphate receptors
Abstract
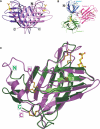
The two members of the P-type lectin family, the 46 kDa cation-dependent mannose 6-phosphate receptor (CD-MPR) and the 300 kDa cation-independent mannose 6-phosphate receptor (CI-MPR), are ubiquitously expressed throughout the animal kingdom and are distinguished from all other lectins by their ability to recognize phosphorylated mannose residues. The best-characterized function of the MPRs is their ability to direct the delivery of approximately 60 different newly synthesized soluble lysosomal enzymes bearing mannose 6-phosphate (Man-6-P) on their N-linked oligosaccharides to the lysosome. In addition to its intracellular role in lysosome biogenesis, the CI-MPR, but not the CD-MPR, participates in a number of other biological processes by interacting with various molecules at the cell surface. The list of extracellular ligands recognized by this multifunctional receptor has grown to include a diverse spectrum of Man-6-P-containing proteins as well as several non-Man-6-P-containing ligands. Recent structural studies have given us a clearer view of how these two receptors use related, but yet distinct, approaches in the recognition of phosphomannosyl residues.
Figures



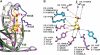
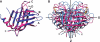
References
-
- Aeed PA, Elhammer AP. Glycosylation of recombinant prorenin in insect cells: The insect cell line Sf9 does not express the mannose 6-phosphate recognition signal. Biochemistry. 1994;33:8793–8797. - PubMed
-
- Bhamidipati A, Denic V, Quan EM, Weissman JS. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol Cell. 2005;19:741–751. - PubMed
-
- Blanchard F, Duplomb L, Raher S, Vusio P, Hoflack B, Jacques Y, Godard A. Mannose 6-phosphate/insulin-like growth factor II receptor mediates internalization and degradation of leukemia inhibitory factor but not signal transduction. J Biol Chem. 1999;274:24685–24693. - PubMed
-
- Blanchard F, Raher S, Duplomb L, Vusio P, Pitard V, Taupin JL, Moreau JF, Hoflack B, Minvielle S, Jacques Y, et al. The mannose 6-phosphate/ insulin-like growth factor II receptor is a nanomolar affinity receptor for glycosylated human leukemia inhibitory factor. J Biol Chem. 1998;273:20886–20893. - PubMed

