Processing of predicted substrates of fungal Kex2 proteinases from Candida albicans, C. glabrata, Saccharomyces cerevisiae and Pichia pastoris
- PMID: 18625069
- PMCID: PMC2515848
- DOI: 10.1186/1471-2180-8-116
Processing of predicted substrates of fungal Kex2 proteinases from Candida albicans, C. glabrata, Saccharomyces cerevisiae and Pichia pastoris
Abstract
Background: Kexin-like proteinases are a subfamily of the subtilisin-like serine proteinases with multiple regulatory functions in eukaryotes. In the yeast Saccharomyces cerevisiae the Kex2 protein is biochemically well investigated, however, with the exception of a few well known proteins such as the alpha-pheromone precursors, killer toxin precursors and aspartic proteinase propeptides, very few substrates are known. Fungal kex2 deletion mutants display pleiotropic phenotypes that are thought to result from the failure to proteolytically activate such substrates.
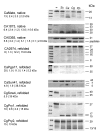
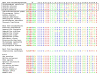
Results: In this study we have aimed at providing an improved assembly of Kex2 target proteins to explain the phenotypes observed in fungal kex2 deletion mutants by in vitro digestion of recombinant substrates from Candida albicans and C. glabrata. We identified CaEce1, CA0365, one member of the Pry protein family and CaOps4-homolog proteins as novel Kex2 substrates.
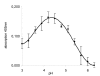
Conclusion: Statistical analysis of the cleavage sites revealed extended subsite recognition of negatively charged residues in the P1', P2' and P4' positions, which is also reflected in construction of the respective binding pockets in the ScKex2 enzyme. Additionally, we provide evidence for the existence of structural constrains in potential substrates prohibiting proteolysis. Furthermore, by using purified Kex2 proteinases from S. cerevisiae, P. pastoris, C. albicans and C. glabrata, we show that while the substrate specificity is generally conserved between organisms, the proteinases are still distinct from each other and are likely to have additional unique substrate recognition.
Figures
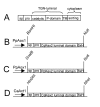
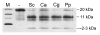
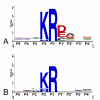

 : Propeptide cleavage site, numbering "a" through "E" refers to residues used in Table 2, which lists the respective binding pockets and references.
: Propeptide cleavage site, numbering "a" through "E" refers to residues used in Table 2, which lists the respective binding pockets and references.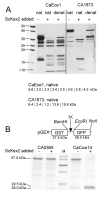

References
-
- Seidah NG, Khatib AM, Prat A. The proprotein convertases and their implication in sterol and/or lipid metabolism. Biol Chem. 2006;387:871–877. - PubMed
-
- Julius D, Brake A, Blair L, Kunisawa R, Thorner J. Isolation of the putative structural gene for the lysine-arginine-cleaving endopeptidase required for processing of yeast prepro-alpha-factor. Cell. 1984;37:1075–1089. - PubMed
-
- Krysan DJ, Rockwell NC, Fuller RS. Quantitative characterization of furin specificity. Energetics of substrate discrimination using an internally consistent set of hexapeptidyl methylcoumarinamides. J Biol Chem. 1999;274:23229–23234. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

