The crystal structure of CHIR-AB1: a primordial avian classical Fc receptor
- PMID: 18625238
- PMCID: PMC2603183
- DOI: 10.1016/j.jmb.2008.06.082
The crystal structure of CHIR-AB1: a primordial avian classical Fc receptor
Abstract
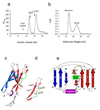
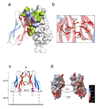
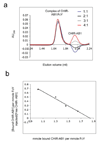
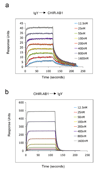
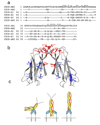
CHIR-AB1 is a newly identified avian immunoglobulin (Ig) receptor that includes both activating and inhibitory motifs and was therefore classified as a potentially bifunctional receptor. Recently, CHIR-AB1 was shown to bind the Fc region of chicken IgY and to induce calcium mobilization via association with the common gamma-chain, a subunit that transmits signals upon ligation of many different immunoreceptors. Here we describe the 1.8-A-resolution crystal structure of the CHIR-AB1 ectodomain. The receptor ectodomain consists of a single C2-type Ig domain resembling the Ig-like domains found in mammalian Fc receptors such as FcgammaRs and FcalphaRI. Unlike these receptors and other monomeric Ig superfamily members, CHIR-AB1 crystallized as a 2-fold symmetrical homodimer that bears no resemblance to variable or constant region dimers in an antibody. Analytical ultracentrifugation demonstrated that CHIR-AB1 exists as a mixture of monomers and dimers in solution, and equilibrium gel filtration revealed a 2:1 receptor/ligand binding stoichiometry. Measurement of the 1:1 CHIR-AB1/IgY interaction affinity indicates a relatively low affinity complex, but a 2:1 CHIR-AB1/IgY interaction allows an increase in apparent affinity due to avidity effects when the receptor is tethered to a surface. Taken together, these results add to the structural understanding of Fc receptors and their functional mechanisms.
Figures
References
-
- Nimmerjahn F, Ravetch JV. Fc-receptors as regulators of immunity. Adv Immunol. 2007;96:179–204. - PubMed
-
- Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol. 2008;8:34–47. - PubMed
-
- Woof JM, Burton DR. Human antibody-Fc receptor interactions illuminated by crystal structures. Nat Rev Immunol. 2004;4:89–99. - PubMed
-
- Garman SC, Kinet JP, Jardetzky TS. Crystal structure of the human high-affinity IgE receptor. Cell. 1998;95:951–961. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

