Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3
- PMID: 18628393
- PMCID: PMC2492733
- DOI: 10.1101/gad.1673508
Control of differentiation in a self-renewing mammalian tissue by the histone demethylase JMJD3
Abstract
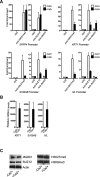
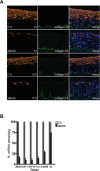
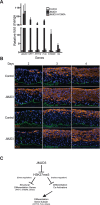
The recent discovery of H3K27me3 demethylases suggests that H3K27me3 may dynamically regulate gene expression, but this potential role in mammalian tissue homeostasis remains uncharacterized. In the epidermis, a tissue that balances stem cell self-renewal with differentiation, H3K27me3, occupies the promoters of many differentiation genes. During calcium-induced differentiation, H3K27me3 was erased at these promoters in concert with loss of PcG protein occupancy and increased binding by the H3K27me3 demethylase, JMJD3. Within epidermal tissue, JMJD3 depletion blocked differentiation, while active JMJD3 dominantly induced it. These results indicate that epigenetic derepression by JMJD3 controls mammalian epidermal differentiation.
Figures
References
-
- Agger K., Cloos P.A., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A.E., Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. - PubMed
-
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Boyce S.T., Ham R.G. Calcium-regulated differentiation of normal human epidermal keratinocytes in chemically defined clonal culture and serum-free serial culture. J. Invest. Dermatol. 1983;81:33s–40s. - PubMed
-
- Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K., et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
