Transcriptional coactivators PGC-1alpha and PGC-lbeta control overlapping programs required for perinatal maturation of the heart
- PMID: 18628400
- PMCID: PMC2492740
- DOI: 10.1101/gad.1661708
Transcriptional coactivators PGC-1alpha and PGC-lbeta control overlapping programs required for perinatal maturation of the heart
Abstract
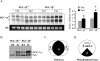
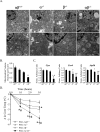
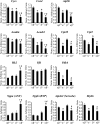
Oxidative tissues such as heart undergo a dramatic perinatal mitochondrial biogenesis to meet the high-energy demands after birth. PPARgamma coactivator-1 (PGC-1) alpha and beta have been implicated in the transcriptional control of cellular energy metabolism. Mice with combined deficiency of PGC-1alpha and PGC-1beta (PGC-1alphabeta(-/-) mice) were generated to investigate the convergence of their functions in vivo. The phenotype of PGC-1beta(-/-) mice was minimal under nonstressed conditions, including normal heart function, similar to that of PGC-1alpha(-/-) mice generated previously. In striking contrast to the singly deficient PGC-1 lines, PGC-1alphabeta(-/-) mice died shortly after birth with small hearts, bradycardia, intermittent heart block, and a markedly reduced cardiac output. Cardiac-specific ablation of the PGC-1beta gene on a PGC-1alpha-deficient background phenocopied the generalized PGC-1alphabeta(-/-) mice. The hearts of the PGC-1alphabeta(-/-) mice exhibited signatures of a maturational defect including reduced growth, a late fetal arrest in mitochondrial biogenesis, and persistence of a fetal pattern of gene expression. Brown adipose tissue (BAT) of PGC-1alphabeta(-/-) mice also exhibited a severe abnormality in function and mitochondrial density. We conclude that PGC-1alpha and PGC-1beta share roles that collectively are necessary for the postnatal metabolic and functional maturation of heart and BAT.
Figures

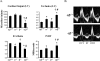


References
-
- Agah R., Frenkel P.A., French B.A., Michael L.H., Overbeek P.A., Schneider M.D. Gene recombination in postmitotic cells. Targeted expression of cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J. Clin. Invest. 1997;100:169–179. - PMC - PubMed
-
- Alaynick W.A., Kondo R.P., Xie W., He W., Dufour C.R., Downes M., Jonker J.W., Giles W., Naviaux R.K., Giguère V., et al. ERRγ directs and maintains the transition to oxidative metabolism in the postnatal heart. Cell Metab. 2007;6:13–24. - PubMed
-
- Arany Z., Lebrasseur N., Morris C., Smith E., Yang W., Ma Y., Chin S., Spiegelman B.M. The transcriptional coactivator PGC-1β drives the formation of oxidative type IIX fibers in skeletal muscle. Cell Metab. 2007;5:35–46. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
