TAF6delta controls apoptosis and gene expression in the absence of p53
- PMID: 18628956
- PMCID: PMC2444026
- DOI: 10.1371/journal.pone.0002721
TAF6delta controls apoptosis and gene expression in the absence of p53
Abstract
Background: Life and death decisions of metazoan cells hinge on the balance between the expression of pro- versus anti-apoptotic gene products. The general RNA polymerase II transcription factor, TFIID, plays a central role in the regulation of gene expression through its core promoter recognition and co-activator functions. The core TFIID subunit TAF6 acts in vitro as an essential co-activator of transcription for the p53 tumor suppressor protein. We previously identified a splice variant of TAF6, termed TAF6delta that can be induced during apoptosis.
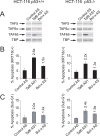
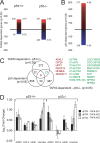
Methodology/principal findings: To elucidate the impact of TAF6delta on cell death and gene expression, we have employed modified antisense oligonucleotides to enforce expression of endogenous TAF6delta. The induction of endogenous TAF6delta triggered apoptosis in tumor cell lines, including cells devoid of p53. Microarray experiments revealed that TAF6delta activates gene expression independently of cellular p53 status.
Conclusions: Our data define TAF6delta as a pivotal node in a signaling pathway that controls gene expression programs and apoptosis in the absence of p53.
Conflict of interest statement
Figures
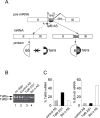
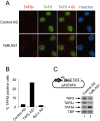
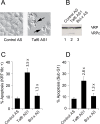
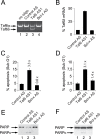
References
-
- Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–776. - PubMed
-
- Thompson CB. Apoptosis in the pathogenesis and treatment of disease. Science. 1995;267:1456–1462. - PubMed
-
- Fesus L. Inducible gene expression in apoptosis [In Process Citation]. Cell Death Differ. 1999;6:1144–1145. - PubMed
-
- Kumar S, Cakouros D. Transcriptional control of the core cell-death machinery. Trends Biochem Sci. 2004;29:193–199. - PubMed
-
- Schwerk C, Schulze-Osthoff K. Regulation of apoptosis by alternative pre-mRNA splicing. Mol Cell. 2005;19:1–13. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

