Global regulation of nucleotide biosynthetic genes by c-Myc
- PMID: 18628958
- PMCID: PMC2444028
- DOI: 10.1371/journal.pone.0002722
Global regulation of nucleotide biosynthetic genes by c-Myc
Abstract
Background: The c-Myc transcription factor is a master regulator and integrates cell proliferation, cell growth and metabolism through activating thousands of target genes. Our identification of direct c-Myc target genes by chromatin immunoprecipitation (ChIP) coupled with pair-end ditag sequencing analysis (ChIP-PET) revealed that nucleotide metabolic genes are enriched among c-Myc targets, but the role of Myc in regulating nucleotide metabolic genes has not been comprehensively delineated.
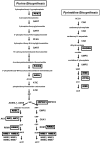
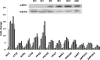
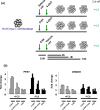
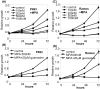
Methodology/principal findings: Here, we report that the majority of genes in human purine and pyrimidine biosynthesis pathway were induced and directly bound by c-Myc in the P493-6 human Burkitt's lymphoma model cell line. The majority of these genes were also responsive to the ligand-activated Myc-estrogen receptor fusion protein, Myc-ER, in a Myc null rat fibroblast cell line, HO.15 MYC-ER. Furthermore, these targets are also responsive to Myc activation in transgenic mouse livers in vivo. To determine the functional significance of c-Myc regulation of nucleotide metabolism, we sought to determine the effect of loss of function of direct Myc targets inosine monophosphate dehydrogenases (IMPDH1 and IMPDH2) on c-Myc-induced cell growth and proliferation. In this regard, we used a specific IMPDH inhibitor mycophenolic acid (MPA) and found that MPA dramatically inhibits c-Myc-induced P493-6 cell proliferation through S-phase arrest and apoptosis.
Conclusions/significance: Taken together, these results demonstrate the direct induction of nucleotide metabolic genes by c-Myc in multiple systems. Our finding of an S-phase arrest in cells with diminished IMPDH activity suggests that nucleotide pool balance is essential for c-Myc's orchestration of DNA replication, such that uncoupling of these two processes create DNA replication stress and apoptosis.
Conflict of interest statement
Figures



References
-
- Hecht JL, Aster JC. Molecular biology of Burkitt's lymphoma. J Clin Oncol. 2000;18:3707–3721. - PubMed
-
- Liao DJ, Dickson RB. c-Myc in breast cancer. Endocr Relat Cancer. 2000;7:143–164. - PubMed
-
- Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. - PubMed
-
- Cory S, Harris AW, Langdon WY, Alexander WS, Corcoran LM, et al. The myc oncogene and lymphoid neoplasia: from translocations to transgenic mice. Hamatol Bluttransfus. 1987;31:248–251. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

