DirecTag: accurate sequence tags from peptide MS/MS through statistical scoring
- PMID: 18630943
- PMCID: PMC2810657
- DOI: 10.1021/pr800154p
DirecTag: accurate sequence tags from peptide MS/MS through statistical scoring
Abstract
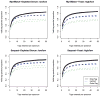
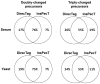
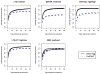
In shotgun proteomics, tandem mass spectra of peptides are typically identified through database search algorithms such as Sequest. We have developed DirecTag, an open-source algorithm to infer partial sequence tags directly from observed fragment ions. This algorithm is unique in its implementation of three separate scoring systems to evaluate each tag on the basis of peak intensity, m/ z fidelity, and complementarity. In data sets from several types of mass spectrometers, DirecTag reproducibly exceeded the accuracy and speed of InsPecT and GutenTag, two previously published algorithms for this purpose. The source code and binaries for DirecTag are available from http://fenchurch.mc.vanderbilt.edu.
Figures




References
-
- Yates JR, III, Eng JK, McCormack AL, Schieltz D. Method to correlate tandem mass spectra of modified peptides to amino acid sequences in the protein database. Anal Chem. 1995;67(8):1426–1436. - PubMed
-
- Sadygov RG, Yates JR., III A hypergeometric probability model for protein identification and validation using tandem mass spectral data and protein sequence databases. Anal Chem. 2003;75(15):3792–3798. - PubMed
-
- Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open mass spectrometry search algorithm. J Proteome Res. 2004;3(5):958–964. - PubMed
-
- Zhang N, Aebersold R, Schwikowski B. ProbID: a probabilistic algorithm to identify peptides through sequence database searching using tandem mass spectral data. Proteomics. 2002;2(10):1406–1412. - PubMed
-
- Fenyo D, Beavis RC. A method for assessing the statistical significance of mass spectrometry-based protein identifications using general scoring schemes. Anal Chem. 2003;75(4):768–774. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

