Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution
- PMID: 18632554
- PMCID: PMC2474566
- DOI: 10.1073/pnas.0800679105
Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution
Abstract
Lateral gene transfer is an important mechanism of natural variation among prokaryotes, but the significance of its quantitative contribution to genome evolution is debated. Here, we report networks that capture both vertical and lateral components of evolutionary history among 539,723 genes distributed across 181 sequenced prokaryotic genomes. Partitioning of these networks by an eigenspectrum analysis identifies community structure in prokaryotic gene-sharing networks, the modules of which do not correspond to a strictly hierarchical prokaryotic classification. Our results indicate that, on average, at least 81 +/- 15% of the genes in each genome studied were involved in lateral gene transfer at some point in their history, even though they can be vertically inherited after acquisition, uncovering a substantial cumulative effect of lateral gene transfer on longer evolutionary time scales.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
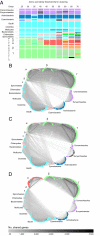
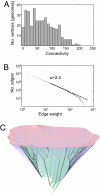
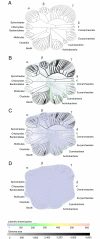
References
-
- Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3:711–721. - PubMed
-
- Lang AS, Beatty JT. Importance of widespread gene transfer agent genes in alpha-proteobacteria. Trends Microbiol. 2007;15:54–62. - PubMed
-
- Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284:2124–2128. - PubMed
-
- Gogarten JP, Doolittle WF, Lawrence JG. Prokaryotic evolution in light of gene transfer. Mol Biol Evol. 2002;19:2226–2238. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

