Crystal structure of coxsackievirus B3 3Dpol highlights the functional importance of residue 5 in picornavirus polymerases
- PMID: 18632862
- PMCID: PMC2546958
- DOI: 10.1128/JVI.00647-08
Crystal structure of coxsackievirus B3 3Dpol highlights the functional importance of residue 5 in picornavirus polymerases
Abstract
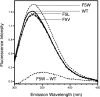
The crystal structure of the coxsackievirus B3 polymerase has been solved at 2.25-A resolution and is shown to be highly homologous to polymerases from poliovirus, rhinovirus, and foot-and-mouth disease viruses. Together, these structures highlight several conserved structural elements in picornaviral polymerases, including a proteolytic activation-dependent N-terminal structure that is essential for full activity. Interestingly, a comparison of all of the picornaviral polymerase structures shows an unusual conformation for residue 5, which is always located at a distortion in the beta-strand composed of residues 1 to 8. In our earlier structure of the poliovirus polymerase, we attributed this conformation to a crystal packing artifact, but the observation that this conformation is conserved among picornaviruses led us to examine the role of this residue in further detail. Here we use coxsackievirus polymerase to show that elongation activity correlates with the hydrophobicity of residue 5 and, surprisingly, more hydrophobic residues result in higher activity. Based on structural analysis, we propose that this residue becomes buried during the nucleotide repositioning step that occurs prior to phosphoryl transfer. We present a model in which the buried N terminus observed in all picornaviral polymerases is essential for stabilizing the structure during this conformational change.
Figures
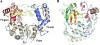


References
-
- Brunger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54(Pt. 5)905-921. - PubMed
-
- DeLano, W. L. 2002. The PYMOL molecular graphics system. DeLano Scientific, San Carlos, CA.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

