Positive feedback of G1 cyclins ensures coherent cell cycle entry
- PMID: 18633409
- PMCID: PMC2606905
- DOI: 10.1038/nature07118
Positive feedback of G1 cyclins ensures coherent cell cycle entry
Abstract
In budding yeast, Saccharomyces cerevisiae, the Start checkpoint integrates multiple internal and external signals into an all-or-none decision to enter the cell cycle. Here we show that Start behaves like a switch due to systems-level feedback in the regulatory network. In contrast to current models proposing a linear cascade of Start activation, transcriptional positive feedback of the G1 cyclins Cln1 and Cln2 induces the near-simultaneous expression of the approximately 200-gene G1/S regulon. Nuclear Cln2 drives coherent regulon expression, whereas cytoplasmic Cln2 drives efficient budding. Cells with the CLN1 and CLN2 genes deleted frequently arrest as unbudded cells, incurring a large fluctuation-induced fitness penalty due to both the lack of cytoplasmic Cln2 and insufficient G1/S regulon expression. Thus, positive-feedback-amplified expression of Cln1 and Cln2 simultaneously drives robust budding and rapid, coherent regulon expression. A similar G1/S regulatory network in mammalian cells, comprised of non-orthologous genes, suggests either conservation of regulatory architecture or convergent evolution.
Figures
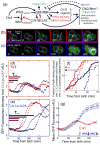

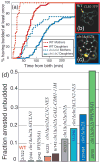

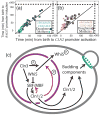
Comment in
-
Systems biology: On the cell cycle and its switches.Nature. 2008 Jul 17;454(7202):288-9. doi: 10.1038/454288a. Nature. 2008. PMID: 18633407 Free PMC article.
References
-
- Simchen G, Pinon R, Salts Y. Sporulation in Saccharomyces cerevisiae: premeiotic DNA synthesis, readiness and commitment. Exp Cell Res. 1972;75:207–18. - PubMed
-
- Nachman I, Regev A, Ramanathan S. Dissecting timing variability in yeast meiosis. Cell. 2007;131:544–56. - PubMed
-
- Ferrell JE, Jr, Machleder EM. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280:895–8. - PubMed
-
- Xiong W, Ferrell JE., Jr A positive-feedback-based bistable ‘memory module’ that governs a cell fate decision. Nature. 2003;426:460–5. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

