Identification of SLEEPLESS, a sleep-promoting factor
- PMID: 18635795
- PMCID: PMC2771549
- DOI: 10.1126/science.1155942
Identification of SLEEPLESS, a sleep-promoting factor
Abstract
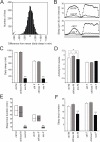
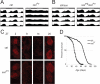
Sleep is an essential process conserved from flies to humans. The importance of sleep is underscored by its tight homeostatic control. Through a forward genetic screen, we identified a gene, sleepless, required for sleep in Drosophila. The sleepless gene encodes a brain-enriched, glycosylphosphatidylinositol-anchored protein. Loss of SLEEPLESS protein caused an extreme (>80%) reduction in sleep; a moderate reduction in SLEEPLESS had minimal effects on baseline sleep but markedly reduced the amount of recovery sleep after sleep deprivation. Genetic and molecular analyses revealed that quiver, a mutation that impairs Shaker-dependent potassium current, is an allele of sleepless. Consistent with this finding, Shaker protein levels were reduced in sleepless mutants. We propose that SLEEPLESS is a signaling molecule that connects sleep drive to lowered membrane excitability.
Figures




Comment in
-
Genetics. Simple sleepers.Science. 2008 Jul 18;321(5887):334-7. doi: 10.1126/science.321.5887.334. Science. 2008. PMID: 18635772 No abstract available.
-
Sleepless in the sea.Science. 2008 Oct 24;322(5901):527. doi: 10.1126/science.322.5901.527a. Science. 2008. PMID: 18948516 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

