Domain-swapped dimerization of ZO-1 PDZ2 generates specific and regulatory connexin43-binding sites
- PMID: 18636092
- PMCID: PMC2516886
- DOI: 10.1038/emboj.2008.138
Domain-swapped dimerization of ZO-1 PDZ2 generates specific and regulatory connexin43-binding sites
Abstract
PDZ domain scaffold proteins are capable of assembling macromolecular protein complexes in diverse cellular processes through PDZ-mediated binding to a short peptide fragment at the carboxyl tail of target proteins. How each PDZ domain specifically recognizes its target protein(s) remains a major conceptual question, as at least a few out of the several hundred PDZ domains in each eukaryotic genome share overlapping binding properties with any given target protein. Here, we show that the domain-swapped dimerization of zonula occludens-1 PDZ2 generates a distinct interface that functions together with the well-separated canonical carboxyl tail-binding pocket in each PDZ unit in binding to connexin43 (Cx43). We further demonstrate that the charge-charge interaction network formed by residues in the PDZ dimer interface and upstream residues of the Cx43 peptide not only provides the unprecedented interaction specificity for the complex but may also function as a phosphorylation-mediated regulatory switch for the dynamics of the Cx43 gap junctions. Finally, we provide evidence that such domain-swapped dimer assembly also occurs in other PDZ domain scaffold proteins. Therefore, our findings present a new paradigm for understanding how some PDZ domain proteins specifically bind to and regulate the functions of their target proteins.
Figures

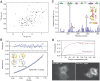
 where ΔδHN and ΔδN represent chemical shift differences of amide proton and nitrogen chemical shifts of the each residue of ZO-1 PDZ2. The scaling factor (αN) used to normalize the 1H and 15N chemical shift is 0.17. (D) Fluorescence-based measurement of the binding affinities of the wild-type ZO-1 PDZ2 domain and the GGGA insertion mutant towards the Cx43 peptide. (E) Comparison of the cellular localizations of the GFP-tagged full-length wild-type ZO-1 and the ZO-1 mutant with the GGGA insertion in its PDZ2 in HeLa cells. The overexpressed wild-type ZO-1 forms plaques at the contact regions between transfected cells, whereas the ZO-1 mutant transfected cells lack such intercellular ZO-1 punta.
where ΔδHN and ΔδN represent chemical shift differences of amide proton and nitrogen chemical shifts of the each residue of ZO-1 PDZ2. The scaling factor (αN) used to normalize the 1H and 15N chemical shift is 0.17. (D) Fluorescence-based measurement of the binding affinities of the wild-type ZO-1 PDZ2 domain and the GGGA insertion mutant towards the Cx43 peptide. (E) Comparison of the cellular localizations of the GFP-tagged full-length wild-type ZO-1 and the ZO-1 mutant with the GGGA insertion in its PDZ2 in HeLa cells. The overexpressed wild-type ZO-1 forms plaques at the contact regions between transfected cells, whereas the ZO-1 mutant transfected cells lack such intercellular ZO-1 punta.

References
-
- Altschuler Y, Hodson C, Milgram SL (2003) The apical compartment: trafficking pathways, regulators and scaffolding proteins. Curr Opin Cell Biol 15: 423–429 - PubMed
-
- Bax A, Grzesiek S (1993) Methodological advances in protein NMR. Acc Chem Res 26: 131–138
-
- Bazzoni G, Martinez-Estrada OM, Orsenigo F, Cordenonsi M, Citi S, Dejana E (2000) Interaction of junctional adhesion molecule with the tight junction components ZO-1, cingulin, and occludin. J Biol Chem 275: 20520–20526 - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

