Molecular mechanisms and diagnosis of chromosome 22q11.2 rearrangements
- PMID: 18636632
- PMCID: PMC2810965
- DOI: 10.1002/ddrr.3
Molecular mechanisms and diagnosis of chromosome 22q11.2 rearrangements
Abstract
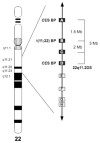
Several recurrent, constitutional genomic disorders are present on chromosome 22q. These include the translocations and deletions associated with DiGeorge and velocardiofacial syndrome and the translocations that give rise to the recurrent t(11;22) supernumerary der(22) syndrome (Emanuel syndrome). The rearrangement breakpoints on 22q cluster around the chromosome-specific segmental duplications of proximal 22q11, which are involved in the etiology of these disorders. While the deletions are the result of nonallelic homologous recombination (NAHR) between low copy repeats or segmental duplications within 22q11, the t(11;22) is the result of rearrangement between palindromic AT-rich repeats on 11q and 22q. Here we describe the mechanisms responsible for these recurrent rearrangements, discuss the recurrent deletion endpoints that are the result of NAHR between chromosome 22q specific low copy repeats as well as present current diagnostic approaches to deletion detection.
Figures
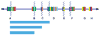


References
-
- Alberti A, Romano C, Falco M, et al. 1.5 Mb de novo 22q11.21 microduplication in a patient with cognitive deficits and dysmorphic facial features. Clin Genet. 2007;71:177–182. - PubMed
-
- Amati F, Conti E, Novelli A, et al. Atypical deletions suggest five 22q11.2 critical regions related to the DiGeorge/velo-cardio-facial syndrome. Eur J Hum Genet. 1999;7:903–909. - PubMed
-
- Baumer A, Dutly F, Balmer D, et al. High level of unequal meiotic crossovers at the origin of the 22q11. 2 and 7q11.23 deletions. Hum Mol Genet. 1998;7:887–894. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

